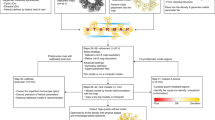
Abstract
Structures of multisubunit macromolecular machines are primarily determined either by electron microscopy (EM) or by X-ray crystallography. In many cases, a structure for a complex can be obtained at low resolution (at a coarse level of detail) with EM and at a higher resolution (with finer detail) by X-ray crystallography. The integration of these two structural techniques is becoming increasingly important for the generation of atomic models of macromolecular complexes. A low-resolution EM image can be a powerful tool for obtaining the 'phase' information that is missing from an X-ray crystallography experiment; however, integration of EM and X-ray diffraction data has been technically challenging. Here we present a step-by-step protocol that explains how low-resolution EM maps can be placed in the crystallographic unit cell by molecular replacement, and how initial phases computed from the placed EM density are extended to high resolution by averaging maps over noncrystallographic symmetry. As the resolution gap between EM and X-ray crystallography continues to narrow, the use of EM maps to help with X-ray crystal structure determination, as described in this protocol, will become increasingly effective.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Garman, E.F. Developments in X-ray crystallographic structure determination of biological macromolecules. Science 343, 1102–1108 (2014).
Rossmann, M.G., Morais, M.C., Leiman, P.G. & Zhang, W. Combining X-ray crystallography and electron microscopy. Structure 13, 355–362 (2005).
Bai, X.-C., McMullan, G. & Scheres, S.H.W. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 40, 49–57 (2014).
Lengyel, J., Hnath, E., Storms, M. & Wohlfarth, T. Towards an integrative structural biology approach: combining Cryo-TEM, X-ray crystallography, and NMR. J. Struct. Funct. Genomics 15, 117–124 (2014).
Rossmann, M.G. Molecular replacement - historical background. Acta Crystallogr. D Biol. Crystallogr. 57, 1360–1366 (2001).
Stuart, D.I. & Abrescia, N.G. From lows to highs: using low-resolution models to phase X-ray data. Acta Crystallogr. D Biol. Crystallogr. 69, 2257–2265 (2013).
Xiong, Y. From electron microscopy to X-ray crystallography: molecular-replacement case studies. Acta Crystallogr. D Biol. Crystallogr. 64, 76–82 (2008).
Dodson, E.J. Using electron-microscopy images as a model for molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 57, 1405–1409 (2001).
Jackson, R.N. et al. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science 345, 1473–1479 (2014).
Mccoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Wiedenheft, B. et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477, 486–489 (2011).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Goddard, T.D., Huang, C.C. & Ferrin, T.E. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure 13, 473–482 (2005).
Kantardjieff, K.A. & Rupp, B. Matthews coefficient probabilities: improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 12, 1865–1871 (2003).
Weichenberger, C.X. & Rupp, B. Ten years of probabilistic estimates of biocrystal solvent content: new insights via nonparametric kernel density estimate. Acta Crystallogr. D Biol. Crystallogr. 70, 1579–1588 (2014).
Zwart, P., Grosse-Kunstleve, R. & Adams, P. Xtriage and Fest: automatic assessment of X-ray data and substructure structure factor estimation. CCP4 Newsletter 43, http://www.ccp4.ac.uk/newsletters/newsletter43/articles/PHZ_RWGK_PDA.pdf (2005).
Zwart, P.H., Grosse-Kunsteleve, R.W. & Adams, P.D. Characterization of X-ray data sets. CCP4 Newsletter 42, https://www.phenix-online.org/papers/ccp4_july_2005_zwart.pdf (2005).
Lowe, J. et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 268, 533–539 (1995).
Rabl, J. et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 30, 360–368 (2008).
Rawat, U.B. et al. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421, 87–90 (2003).
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006).
Acknowledgements
R.N.J. is supported by the National Research Service Award postdoctoral fellowship (F32 GM108436) from the US National Institutes of Health (NIH). R.J.R. and T.C.T. are supported by a grant (GM063210) from the NIH. R.J.R. is supported by a Principal Research Fellowship from the Wellcome Trust (grant no. 082961/Z/07/Z). Research in the Wiedenheft lab is supported by the NIH IDeA Program COBRE (GM110732), an R01 to B.W. (GM108888), the National Science Foundation EPSCoR (EPS-110134), the M.J. Murdock Charitable Trust and the Montana State University Agricultural Experimental Station.
Author information
Authors and Affiliations
Contributions
All authors contributed to writing the protocol. A.J.M. developed the EM refinement parameter in Phaser-MR necessary to account for uncertainty in the EM magnification error.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note (PDF 95 kb)
Supplementary Data 1–13
Zip file containing Supplementary Data (ZIP 50241 kb)
Rights and permissions
About this article
Cite this article
Jackson, R., McCoy, A., Terwilliger, T. et al. X-ray structure determination using low-resolution electron microscopy maps for molecular replacement. Nat Protoc 10, 1275–1284 (2015). https://doi.org/10.1038/nprot.2015.069
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.069
This article is cited by
-
Progerin impairs 3D genome organization and induces fragile telomeres by limiting the dNTP pools
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.