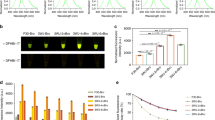
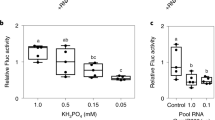
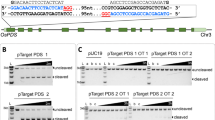
Abstract
Artificial miRNA (amiRNA) technology offers highly specific gene silencing in diverse plant species. The principal challenge in amiRNA application is to select potent amiRNAs from hundreds of bioinformatically designed candidates to enable maximal target gene silencing at the protein level. To address this issue, we developed the epitope-tagged protein-based amiRNA (ETPamir) screens, in which single or multiple potential target genes encoding epitope-tagged proteins are constitutively or inducibly coexpressed with individual amiRNA candidates in plant protoplasts. Accumulation of tagged proteins, detected by immunoblotting with commercial tag antibodies, inversely and quantitatively reflects amiRNA efficacy in vivo. The core procedure, from protoplast isolation to identification of optimal amiRNA, can be completed in 2–3 d. The ETPamir screens circumvent the limited availability of plant antibodies and the complexity of plant amiRNA silencing at target mRNA and/or protein levels. The method can be extended to verify predicted target genes for endogenous plant miRNAs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Zhang, F. et al. High-frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA 107, 12028–12033 (2010).
Li, T., Liu, B., Spalding, M.H., Weeks, D.P. & Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392 (2012).
Christian, M., Qi, Y., Zhang, Y. & Voytas, D.F. Targeted mutagenesis of Arabidopsis thaliana using engineered TAL effector nucleases. G3 3, 1697–1705 (2013).
Li, J.F. et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691 (2013).
Shan, Q. et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688 (2013).
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J.D.G. & Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693 (2013).
Wang, Y. et al. Modes of gene duplication contribute differently to genetic novelty and redundancy but show parallels across divergent angiosperms. PLoS ONE 6, e28150 (2011).
Alvarez, J.P. et al. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell 18, 1134–1151 (2006).
Schwab, R., Ossowski, S., Riester, M., Warthmann, N. & Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133 (2006).
Ossowski, S., Schwab, R. & Weigel, D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53, 674–690 (2008).
Li, J.F. et al. Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. Plant Cell 25, 1507–1522 (2013).
Parizotto, E.A., Dunoyer, P., Rahm, N., Himber, C. & Voinnet, O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18, 2237–2242 (2004).
Niu, Q.W. et al. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 24, 1420–1428 (2006).
Eamens, A.L., McHale, M. & Waterhouse, P.M. The use of artificial microRNA technology to control gene expression in Arabidopsis thaliana. Methods Mol. Biol. 1062, 211–224 (2014).
Warthmann, N., Chen, H., Ossowski, S., Weigel, D. & Herve, P. Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE 3, e1829 (2008).
Park, W., Zhai, J. & Lee, J.Y. Highly efficient gene silencing using perfect complementary artificial miRNA targeting AP1 or heteromeric artificial miRNA targeting AP1 and CAL genes. Plant Cell Rep. 28, 469–480 (2009).
Deveson, I., Li, J. & Millar, A.A. MicroRNAs with analogous target complementarities perform with highly variable efficacies in Arabidopsis. FEBS Lett. 587, 3703–3708 (2013).
Bhagwat, B. et al. An in vivo transient expression system can be applied for rapid and effective selection of artificial microRNA constructs for plant stable genetic transformation. J. Genet. Genomics 40, 261–270 (2013).
Pasquinelli, A.E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13, 271–282 (2012).
Yoo, S.D., Cho, Y.H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Felippes, F.F., Wang, J.W. & Weigel, D. MIGS: miRNA-induced gene silencing. Plant J. 70, 541–547 (2012).
Weiberg, A. et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013).
Llave, C., Xie, Z., Kasschau, K.D. & Carrington, J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297, 2053–2056 (2002).
Brodersen, P. et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190 (2008).
Iwakawa, H.O. & Tomari, Y. Molecular insights into microRNA-mediated translational repression in plants. Mol. Cell 52, 591–601 (2013).
Palatnik, J.F. et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev. Cell 13, 115–125 (2007).
Li, J.F., Li, L. & Sheen, J. A rapid and economical procedure for purification of plasmid or plant DNA with diverse applications in plant biology. Plant Methods 6, 1 (2010).
Gursanscky, N.R., Searle, I.R. & Carroll, B.J. Mobile microRNAs hit the target. Traffic 12, 1475–1482.
Oliveira, M.M., Barroso, J. & Pais, M.S. Direct gene transfer into Actinidia deliciosa protoplasts: analysis of transient expression of the CAT gene using TLC autoradiography and a GC-MS-based method. Plant Mol. Biol. 17, 235–242 (1991).
Li, Z., Cheng, M., Demski, J.W. & Jarret, R.L. Improved electroporation buffer enhances transient gene expression in Arachis hypogaea protoplasts. Genome 38, 858–863 (1995).
Huttly, A.K. & Baulcombe, D.C. A wheat α-Amy2 promoter is regulated by gibberellin in transformed oat aleurone protoplasts. EMBO J. 8, 1907–1913 (1989).
Pauls, P.K., Kunert, K., Huttner, E. & Grandbastien, M.A. Expression of the tobacco Tnt1 retrotransposon promoter in heterologous species. Plant Mol. Biol. 26, 393–402 (1994).
Eimert, K. & Siegemund, F. Transformation of cauliflower (Brassica oleracea L. var. botrytis)-an experimental survey. Plant Mol. Biol. 19, 485–490 (1992).
Chung, E. et al. Molecular and biochemical characterization of the Capsicum annuum calcium-dependent protein kinase 3 (CaCDPK3) gene induced by abiotic and biotic stresses. Planta 220, 286–295 (2004).
Jiang, L. & Pan, L.J. Identification and expression of C2H2 transcription factor genes in Carica papaya under abiotic and biotic stresses. Mol. Biol. Rep. 39, 7105–7115 (2012).
Kindle, K.L. High-efficiency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 87, 1228–1232 (1990).
Niedz, R.P., McKendree, W.L. & Shatters Jr, R.C. Electroporation of embryogenic protoplasts of sweet orange (Citrus sinensis L. Osbeck) and regeneration of transformed plants. In Vitro Cell. Dev. Biol. –Plant 39, 586–594 (2003).
Graham, I.A., Baker, C.J. & Leaver, C.J. Analysis of the cucumber malate synthase gene promoter by transient expression and gel retardation assays. Plant J. 6, 893–902 (1994).
Wang, Z.Y. et al. Transgenic plants of tall fescue (Festuca arundinacea Schreb.) obtained by direct gene transfer to protoplasts. Biotechnology 10, 691–696 (1992).
Lin, W., Odell, J.T. & Schreiner, R.M. Soybean protoplast culture and direct gene uptake and expression by cultured soybean protoplasts. Plant Physiol. 84, 856–861 (1987).
Gao, X. et al. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305 (2011).
Gopalakrishnan, B., Sonthayanon, B., Rahmatullah, R. & Muthukrishnan, S. Barley aleurone layer cell protoplasts as a transient expression system. Plant Mol. Biol. 16, 463–467 (1991).
Czarnecka, E., Verner, F.L. & Gurley, W.B. A strategy for building an amplified transcriptional switch to detect bacterial contamination of plants. Plant Mol. Biol. 78, 59–75 (2012).
Loake, G.J. et al. Phenylpropanoid pathway intermediates regulate transient expression of a chalcone synthase gene promoter. Plant Cell 3, 829–840 (1991).
Kobayashi, Y., Dokiya, Y., Sugiura, M., Niwa, Y. & Sugita, M. Genomic organization and organ-specific expression of a nuclear gene encoding phage-type RNA polymerase in Nicotiana sylvestris. Gene 279, 33–40 (2001).
Ranjan, R. et al. Development and functional analysis of novel genetic promoters using DNA shuffling, hybridization and a combination thereof. PLoS ONE 7, e31931 (2012).
Zhang, Y. et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7, 30 (2011).
Mazarei, M., Al-Ahmad, H., Rudis, M.R. & Stewart Jr, C.N. Protoplast isolation and transient gene expression in switchgrass, Panicum virgatum L. Biotechnol. J. 3, 354–359 (2008).
de Lange, P., de Boer, G.J., Mol, J.N. & Kooter, J.M. Conditional inhibition of β-glucuronidase expression by antisense gene fragments in petunia protoplasts. Plant Mol. Biol. 23, 45–55 (1993).
Roby, D., Broglie, K., Gaynor, J. & Broglie, R. Regulation of a chitinase gene promoter by ethylene and elicitors in bean protoplasts. Plant Physiol. 97, 433–439 (1991).
Thevenin, J. et al. A new system for fast and quantitative analysis of heterologous gene expression in plants. New Phytol. 193, 504–512 (2012).
Gomez-Maldonado, J., Crespillo, R., Avila, C. & Canovas, F.M. Efficient preparation of maritime pine (Pinus pinaster) protoplasts suitable for transgene expression analysis. Plant Mol. Biol. Rep. 19, 361–366 (2001).
Ballas, N., Wong, L.M. & Theologis, A. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum). J. Mol. Biol. 233, 580–596 (1993).
Guo, J. et al. Highly efficient isolation of Populus mesophyll protoplasts and its application in transient expression assays. PLoS ONE 7, e44908 (2012).
Gao, S.J. et al. Enhanced transgene expression in sugarcane by co-expression of virus-encoded RNA silencing suppressors. PLoS ONE 8, e66046 (2013).
Yin, C., Richter, U., Borner, T. & Weihe, A. Evolution of phage-type RNA polymerases in higher plants: characterization of the single phage-type RNA polymerase gene from Selaginella moellendorffii. J. Mol. Evol. 68, 528–538 (2009).
Feltkamp, D., Masterson, R., Starke, J. & Rosahl, S. Analysis of the involvement of ocs-like bZip-binding elements in the differential strength of the bidirectional mas1′2′ promoter. Plant Physiol. 105, 259–268 (1994).
Wahler, D. et al. Polyphenoloxidase silencing affects latex coagulation in Taraxacum species. Plant Physiol. 151, 334–346 (2009).
Lee, B., Murdoch, K., Topping, J., Kreis, M. & Jones, M.G. Transient gene expression in aleurone protoplasts isolated from developing caryopses of barley and wheat. Plant Mol. Biol. 13, 21–29 (1989).
Mirabella, R., Franken, C., van der Krogt, G.N., Bisseling, T. & Geurts, R. Use of the fluorescent timer DsRED-E5 as reporter to monitor dynamics of gene activity in plants. Plant Physiol. 135, 1879–1887 (2004).
Marchive, C. et al. Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PLoS ONE 8, e54185 (2013).
Sheen, J. Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. Plant Cell 3, 225–245 (1991).
Liu, C. et al. A simple artificial microRNA vector based on ath-miR169d precursor from Arabidopsis. Mol. Biol. Rep. 37, 256–262 (2009).
Zhao, T., Wang, W., Bai, X. & Qi, Y. Gene silencing by artificial microRNAs in Chlamydomonas. Plant J. 58, 157–164 (2009).
Molnar, A. et al. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 58, 165–174 (2009).
Melito, S. et al. A nematode demographics assay in transgenic roots reveals no significant impacts of the Rhg1 locus LRR-Kinase on soybean cyst nematode resistance. BMC Plant Biol. 10, 104 (2010).
Ali, I., Amin, I., Briddon, R.W. & Mansoor, S. Artificial microRNA-mediated resistance against the monopartite begomovirus Cotton leaf curl Burewala virus. Virol. J. 10, 231 (2013).
Verdonk, J.C. & Sullivan, M.L. Artificial microRNA (amiRNA) induced gene silencing in alfalfa (Medicago sativa). Botany 91, 117–122 (2013).
Devers, E.A., Teply, J., Reinert, A., Gaude, N. & Krajinski, F. An endogenous artificial microRNA system for unraveling the function of root endosymbioses related genes in Medicago truncatula. BMC Plant Biol. 13, 82 (2013).
Khraiwesh, B., Ossowski, S., Weigel, D., Reski, R. & Frank, W. Specific gene silencing by artificial microRNAs in Physcomitrella patens: an alternative to targeted gene knockouts. Plant Physiol. 148, 684–693 (2008).
Shi, R., Yang, C., Lu, S., Sederoff, R. & Chiang, V.L. Specific down-regulation of PAL genes by artificial microRNAs in Populus trichocarpa. Planta 232, 1281–1288 (2010).
Fernandez, A.I. et al. Flexible tools for gene expression and silencing in tomato. Plant Physiol. 151, 1729–1740 (2009).
Toppino, L. et al. Reversible male sterility in eggplant (Solanum melongena L.) by artificial microRNA-mediated silencing of general transcription factor genes. Plant Biotechnol. J. 9, 684–692 (2011).
Fahim, M., Millar, A.A., Wood, C.C. & Larkin, P.J. Resistance to wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat. Plant Biotechnol. J. 10, 150–163 (2012).
Roumi, V. et al. Transient expression of artificial microRNAs confers resistance to grapevine virus A in Nicotiana benthamiana. J. Plant Pathol. 94, 643–649 (2012).
Debernardi, J.M., Rodriguez, R.E., Mecchia, M.A. & Palatnik, J.F. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 8, e1002419 (2012).
Acknowledgements
We thank the Weigel laboratory for developing the versatile WMD platform and members in the Sheen laboratory for their efforts to test and improve the ETPamir screens. This work has been supported by a Massachusetts General Hospital Executive Committee on Research Postdoctoral Fellowship for Medical Discovery to J.-F.L. and by grants from the US National Science Foundation (grant no. IOS-0843244) and the US National Institutes of Health (grant nos. R01 GM60493 and R01 GM70567) to J.S.
Author information
Authors and Affiliations
Contributions
J.-F.L. developed the protocol under the guidance of J.S. The article was written by J.-F.L. and J.S. D.Z. prepared Tables 1 and 2 and related references.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Li, JF., Zhang, D. & Sheen, J. Epitope-tagged protein-based artificial miRNA screens for optimized gene silencing in plants. Nat Protoc 9, 939–949 (2014). https://doi.org/10.1038/nprot.2014.061
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.061
This article is cited by
-
Designing Climate-Resilient Crops for Sustainable Agriculture: A Silent Approach
Journal of Plant Growth Regulation (2023)
-
Integrated transcriptomic and metabolomic analysis provides insight into the regulation of leaf senescence in rice
Scientific Reports (2021)
-
BRAHMA-interacting proteins BRIP1 and BRIP2 are core subunits of Arabidopsis SWI/SNF complexes
Nature Plants (2020)
-
Ligand-induced monoubiquitination of BIK1 regulates plant immunity
Nature (2020)
-
Silencing AC1 of Tomato leaf curl virus using artificial microRNA confers resistance to leaf curl disease in transgenic tomato
Plant Cell Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.