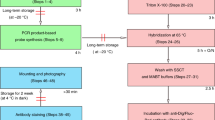
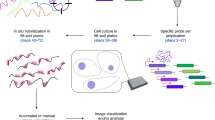
Abstract
RNA in situ hybridization (ISH) has been widely used in cell and developmental biology research to study gene expression. Classical ISH protocols use colorimetric staining approaches, such as the assay with nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate (NBT/BCIP), which do not allow the implementation of multiple probe analyses and do not enable investigators to achieve cellular resolution. Here we describe a protocol to determine the presence of target cytoplasmic RNA via cytoplasmic fluorescence ISH (cFISH), an approach that renders possible the visualization of specific RNA strands from the whole tissue down to the cell. This fluorescence technique, adapted here for use in mouse embryos, enables researchers to implement multiple labeling by combining several RNA probes and/or antibodies in immuno-cFISH. Depending on the options chosen, the protocol can be completed within 2 or 3 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cox, K.H., DeLeon, D.V., Angerer, L.M. & Angerer, R.C. Detection of mrnas in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev. Biol. 101, 485–502 (1984).
Ingham, P.W., Howard, K.R. & Ish-Horowicz, D. Transcription pattern of the Drosophila segmentation gene hairy. Nature 318, 439–445 (1985).
Tautz, D. & Pfeifle, C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98, 81–85 (1989).
Rosen, B. & Beddington, R.S. Whole-mount in situ hybridization in the mouse embryo: gene expression in three dimensions. Trends Genet. 9, 162–167 (1993).
Wijgerde, M., Grosveld, F. & Fraser, P. Transcription complex stability and chromatin dynamics in vivo. Nature 377, 209–213 (1995).
Namekawa, S.H. & Lee, J.T. Detection of nascent RNA, single-copy DNA and protein localization by immunoFISH in mouse germ cells and preimplantation embryos. Nat. Protoc. 6, 270–284 (2011).
Chazaud, C., Yamanaka, Y., Pawson, T. & Rossant, J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev. Cell 10, 615–624 (2006).
Chazaud, C. & Rossant, J. Disruption of early proximodistal patterning and AVE formation in Apc mutants. Development 133, 3379–3387 (2006).
Hauptmann, G. & Gerster, T. Two-color whole-mount in situ hybridization to vertebrate and Drosophila embryos. Trends Genet. 10, 266 (1994).
Sarret, C. et al. Novel neuronal proteolipid protein isoforms encoded by the human myelin proteolipid protein 1 gene. Neuroscience 166, 522–538 (2010).
Nagy, A., Gertsenstein, M., Vintersten, K. & Behringer, K. Manipulating The Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2003).
Sambrook, J. & Russell, D.W. Dot and slot hybridization of purified RNA. Cold Spring Harb. Protoc. doi:10.1101/pdb.prot4052 (2006).
Vize, P.D., McCoy, K.E. & Zhou, X. Multichannel wholemount fluorescent and fluorescent/chromogenic in situ hybridization in Xenopus embryos. Nat. Protoc. 4, 975–983 (2009).
Acknowledgements
These protocols were originally developed by C.C. in J. Rossant's laboratory, on the basis of a chromogenic protocol13. We thank B. Ciruna for the initial use of tyramide amplification. This work has been supported by the 'Ligue Nationale contre le Cancer- CD 63' and by the Agence Nationale de la Recherche (ANR) 'EpiNodal'. C.D. was supported with a fellowship from the ANR 'EpiNodal'. The adult and fetal human tissues were obtained from the Brain and Tissue Bank for Developmental Disorders at the University of Maryland.
Author information
Authors and Affiliations
Contributions
M.G., C.D. and C.C. wrote the manuscript; C.C. designed the protocols; M.G. and C.D. performed the experiments on mouse embryos and C.V.-B. on human brain tissues; and C.C. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Gasnier, M., Dennis, C., Vaurs-Barrière, C. et al. Fluorescent mRNA labeling through cytoplasmic FISH. Nat Protoc 8, 2538–2547 (2013). https://doi.org/10.1038/nprot.2013.160
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.160
This article is cited by
-
Incorporation of isotopic, fluorescent, and heavy-atom-modified nucleotides into RNAs by position-selective labeling of RNA
Nature Protocols (2018)
-
Cripto is essential to capture mouse epiblast stem cell and human embryonic stem cell pluripotency
Nature Communications (2016)
-
A close look at the mammalian blastocyst: epiblast and primitive endoderm formation
Cellular and Molecular Life Sciences (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.