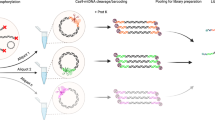
Abstract
Parallel tagged sequencing (PTS) is a molecular barcoding method designed to adapt the recently developed high-throughput 454 parallel sequencing technology for use with multiple samples. Unlike other barcoding methods, PTS can be applied to any type of double-stranded DNA (dsDNA) sample, including shotgun DNA libraries and pools of PCR products, and requires no amplification or gel purification steps. The method relies on attaching sample-specific barcoding adapters, which include sequence tags and a restriction site, to blunt-end repaired DNA samples by ligation and strand-displacement. After pooling multiple barcoded samples, molecules without sequence tags are effectively excluded from sequencing by dephosphorylation and restriction digestion, and using the tag sequences, the source of each DNA sequence can be traced. This protocol allows for sequencing 300 or more complete mitochondrial genomes on a single 454 GS FLX run, or twenty-five 6-kb plasmid sequences on only one 16th plate region. Most of the reactions can be performed in a multichannel setup on 96-well reaction plates, allowing for processing up to several hundreds of samples in a few days.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sanger, F., Nicklen, S. & Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA. 74, 5463–5467 (1977).
Hutchison, C.A. 3rd. DNA sequencing: bench to bedside and beyond. Nucleic Acids Res. 35, 6227–6237 (2007).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Bentley, D.R. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 16, 545–552 (2006).
Thomas, R.K. et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picolitre reactor sequencing. Nat. Med. 12, 852–855 (2006).
Poinar, H.N. et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science 311, 392–394 (2006).
Hofreuter, D. et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74, 4694–4707 (2006).
Gowda, M. et al. Robust analysis of 5′-transcript ends (5′-RATE): a novel technique for transcriptome analysis and genome annotation. Nucleic Acids Res. 34, e126 (2006).
Tawfik, D.S. & Griffiths, A.D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 16, 652–656 (1998).
Meyer, M., Stenzel, U., Myles, S., Prufer, K. & Hofreiter, M. Targeted high-throughput sequencing of tagged nucleic acid samples. Nucleic Acids Res. 35, e97 (2007).
Simcox, T.G. et al. SrfI, a new type-II restriction endonuclease that recognizes the octanucleotide sequence, (sequence: see text). Gene 109, 121–123 (1991).
Pääbo, S. et al. Genetic analyses from ancient DNA. Annu. Rev. Genet. 38, 645–679 (2004).
Willerslev, E. & Cooper, A. Ancient DNA. Proc. Biol. Sci. 272, 3–16 (2005).
Roca, A.L., Georgiadis, N., Pecon-Slattery, J. & O'Brien, S.J. Genetic evidence for two species of elephant in Africa. Science 293, 1473–1477 (2001).
Lindblad-Toh, K. et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 (2005).
Johnson, W.E. et al. The late Miocene radiation of modern Felidae: a genetic assessment. Science 311, 73–77 (2006).
Nielsen, K.L., Hogh, A.L. & Emmersen, J. DeepSAGE—digital transcriptomics with high sensitivity, simple experimental protocol and multiplexing of samples. Nucleic Acids Res. 34, e133 (2006).
Binladen, J. et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PloS ONE 2, e197 (2007).
Hoffmann, C. et al. DNA bar coding and pyrosequencing to identify rare HIV drug resistance mutations. Nucleic Acids Res. 35, e91 (2007).
Parameswaran, P. et al. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res. 35, e130 (2007).
Moore, M.J. et al. Rapid and accurate pyrosequencing of angiosperm plastid genomes. BMC Plant Biol. 6, 17 (2006).
Wicker, T. et al. 454 sequencing put to the test using the complex genome of barley. BMC Genomics 7, 275 (2006).
Huse, S.M., Huber, J.A., Morrison, H.G., Sogin, M.L. & Welch, D.M. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8, R143 (2007).
DeAngelis, M.M., Wang, D.G. & Hawkins, T.L. Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res. 23, 4742–4743 (1995).
Meyer, M. et al. From micrograms to picograms: quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res. 15 Dec. 2007 (Epub ahead of print).
Ahn, S.J., Costa, J. & Emanuel, J.R. PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res. 24, 2623–2625 (1996).
Singer, V.L., Jones, L.J., Yue, S.T. & Haugland, R.P. Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal. Biochem. 249, 228–238 (1997).
Acknowledgements
We thank Christine Green for comments on the manuscript, Ellen Gunnarsdottir for help with providing data and Knut Finstermeier for help with the figures. This work was funded by the Max Planck Society.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Table 1
Tags for designing barcoding oligos. Listed are 6-, 7- and 8-nt tags with a minimum distance of two or three substitutions. See Fig. 2b for instructions on how to convert the tags into full oligo sequences. (DOC 38 kb)
Rights and permissions
About this article
Cite this article
Meyer, M., Stenzel, U. & Hofreiter, M. Parallel tagged sequencing on the 454 platform. Nat Protoc 3, 267–278 (2008). https://doi.org/10.1038/nprot.2007.520
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.520
This article is cited by
-
The value of twinned pollinator-pollen metabarcoding: bumblebee pollination service is weakly partitioned within a UK grassland community
Scientific Reports (2023)
-
Robust and scalable barcoding for massively parallel long-read sequencing
Scientific Reports (2022)
-
Biosurfactant-Producing Capability and Prediction of Functional Genes Potentially Beneficial to Microbial Enhanced Oil Recovery in Indigenous Bacterial Communities of an Onshore Oil Reservoir
Current Microbiology (2019)
-
Association genetics of essential oil traits in Eucalyptus loxophleba: explaining variation in oil yield
Molecular Breeding (2017)
-
Biodegradation of high concentrations of mixed polycyclic aromatic hydrocarbons by indigenous bacteria from a river sediment: a microcosm study and bacterial community analysis
Environmental Science and Pollution Research (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.