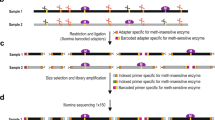
Abstract
Pyrosequencing is a sequencing-by-synthesis method that quantitatively monitors the real-time incorporation of nucleotides through the enzymatic conversion of released pyrophosphate into a proportional light signal. Quantitative measures are of special importance for DNA methylation analysis in various developmental and pathological situations. Analysis of DNA methylation patterns by pyrosequencing combines a simple reaction protocol with reproducible and accurate measures of the degree of methylation at several CpGs in close proximity with high quantitative resolution. After bisulfite treatment and PCR, the degree of each methylation at each CpG position in a sequence is determined from the ratio of T and C. The process of purification and sequencing can be repeated for the same template to analyze other CpGs in the same amplification product. Quantitative epigenotypes are obtained using this protocol in approximately 4 h for up to 96 DNA samples when bisulfite-treated DNA is already available as the starting material.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ronaghi, M., Karamohamed, S., Pettersson, B., Uhlén, M. & Nyrén, P. Real-time DNA sequencing using detection of pyrophosphate release. Anal. Biochem. 242, 84–89 (1996).
Ronaghi, M., Uhlén, M. & Nyrén, P. A sequencing method based on real-time pyrophosphate. Science 281 363, 365 (1998).
Langaee, T. & Ronaghi, M. Genetic variation analyses by Pyrosequencing. Mutat. Res. 573, 96–102 (2005).
Ogino, S. et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J. Mol. Diagn. 7, 413–421 (2005).
Clarke, S.C. Pyrosequencing: nucleotide sequencing technology with bacterial genotyping applications. Expert Rev. Mol. Diagn. 5, 947–953 (2005).
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Rickert, A.M., Premstaller, A., Gebhardt, C. & Oefner, P.J. Genotyping of Snps in a polyploid genome by pyrosequencing. Biotechniques 32, 592–593, 596–598, 600 passim (2002).
Gruber, J.D., Colligan, P.B. & Wolford, J.K. Estimation of single nucleotide polymorphism allele frequency in DNA pools by using pyrosequencing. Hum. Genet. 110, 395–401 (2002).
Lavebratt, C. & Sengul, S. Single nucleotide polymorphism (SNP) allele frequency estimation in DNA pools using pyrosequencing. Nat. Protoc. 1, 2573–2582 (2006).
Pielberg, G., Day, A.E., Plastow, G.S. & Andersson, L. A sensitive method for detecting variation in copy numbers of duplicated genes. Genome Res. 13, 2171–2177 (2003).
Deutsch, S. et al. Detection of aneuploidies by paralogous sequence quantification. J. Med. Genet. 41, 908–915 (2004).
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002).
Jones, P.A. & Baylin, S.B. The epigenomics of cancer. Cell 128, 683–692 (2007).
Laird, P.W. Early detection: the power and the promise of DNA methylation markers. Nat. Rev. Cancer 3, 253–266 (2003).
Brena, R.M., Huang, T.H. & Plass, C. Quantitative assessment of DNA methylation: potential applications for disease diagnosis, classification, and prognosis in clinical settings. J. Mol. Med. 84, 365–377 (2006).
Ehrich, M. et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. USA 102, 15785–15790 (2005).
Colella, S., Shen, L., Baggerly, K.A., Issa, J.P. & Krahe, R. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques 35, 146–150 (2003).
Tost, J., Dunker, J. & Gut, I.G. Analysis and quantification of multiple methylation variable positions in CpG islands by pyrosequencing. Biotechniques 35, 152–156 (2003).
Uhlmann, K., Brinckmann, A., Toliat, M.R., Ritter, H. & Nürnberg, P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis 23, 4072–4079 (2002).
Tost, J., El Abdalaoui, H. & Gut, I.G. Serial pyrosequencing for quantitative DNA methylation analysis. Biotechniques 40, 721–722, 724, 726 (2006).
Mirmohammadsadegh, A. et al. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 66, 6546–6552 (2006).
Xinarianos, G. et al. Frequent genetic and epigenetic abnormalities contribute to the deregulation of cytoglobin in non-small cell lung cancer. Hum. Mol. Genet. 15, 2038–2044 (2006).
Schatz, P., Dietrich, D. & Schuster, M. Rapid analysis of CpG methylation patterns using RNase T1 cleavage and MALDI-TOF. Nucleic Acids Res. 32, e167 (2004).
Yang, A.S. et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 66, 5495–5503 (2006).
White, H.E., Durston, V.J., Harvey, J.F. & Cross, N.C. Quantitative analysis of SNRPN (correction of SNRPN) gene methylation by pyrosequencing as a diagnostic test for Prader-Willi syndrome and Angelman syndrome. Clin. Chem. 52, 1005–1013 (2006).
Wong, H.L. et al. Rapid and quantitative method of allele-specific DNA methylation analysis. Biotechniques 41, 734–739 (2006).
Yang, A.S. et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 32, e38 (2004).
Karimi, M. et al. LUMA (LUminometric Methylation Assay)—a high throughput method to the analysis of genomic DNA methylation. Exp. Cell Res. 312, 1989–1995 (2006).
Li, L.C. & Dahiya, R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 18, 1427–1431 (2002).
Arányi, T., Váradi, A., Simon, I. & Tusnády, G.E. The BiSearch web server. BMC Bioinformatics 7, 431 (2006).
Olek, A., Oswald, J. & Walter, J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 24, 5064–5066 (1996).
Boyd, V.L. & Zon, G. Bisulfite conversion of genomic DNA for methylation analysis: protocol simplification with higher recovery applicable to limited samples and increased throughput. Anal. Biochem. 326, 278–280 (2004).
Bian, Y.S., Yan, P., Osterheld, M.C., Fontolliet, C. & Benhattar, J. Promoter methylation analysis on microdissected paraffin-embedded tissues using bisulfite treatment and PCR-SSCP. Biotechniques 30, 66–72 (2001).
Kerjean, A. et al. Bisulfite genomic sequencing of microdissected cells. Nucleic Acids Res. 29, e106 (2001).
Shiraishi, M. & Hayatsu, H. High-speed conversion of cytosine to uracil in bisulfite genomic sequencing analysis of DNA methylation. DNA Res. 11, 409–415 (2004).
Dupont, J.M., Tost, J., Jammes, H. & Gut, I.G. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal. Biochem. 333, 119–127 (2004).
Warnecke, P.M. et al. Detection and measurement of PCR bias in quantitative methylation analysis of bisulphite-treated DNA. Nucleic Acids Res. 25, 4422–4426 (1997).
Wojdacz, T.K. & Hansen, L.L. Reversal of PCR bias for improved sensitivity of the DNA methylation melting curve assay. Biotechniques 41 274, 276, 278 (2006).
Shen, L., Guo, Y., Chen, X., Ahmed, S. & Issa, J.P. Optimizing annealing temperature overcomes bias in bisulfite PCR methylation analysis. Biotechniques 42, 48–52 (2007).
Acknowledgements
This work was supported by the French Ministry of Research and the European Commission under the Integrated Project 'MolPage' (contract number LSHG-CT-2004-512966).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Tost, J., Gut, I. DNA methylation analysis by pyrosequencing. Nat Protoc 2, 2265–2275 (2007). https://doi.org/10.1038/nprot.2007.314
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.314
This article is cited by
-
Unlocking the secrets: the power of methylation-based cfDNA detection of tissue damage in organ systems
Clinical Epigenetics (2023)
-
Genome-wide DNA methylation analysis in schizophrenia with tardive dyskinesia: a preliminary study
Genes & Genomics (2023)
-
Epigenetically regulated PCDHB15 impairs aggressiveness of metastatic melanoma cells
Clinical Epigenetics (2022)
-
Enzyme-free targeted DNA demethylation using CRISPR–dCas9-based steric hindrance to identify DNA methylation marks causal to altered gene expression
Nature Protocols (2022)
-
Prepubertal nutritional modulation in the bull and its impact on sperm DNA methylation
Cell and Tissue Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.