Abstract
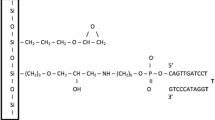
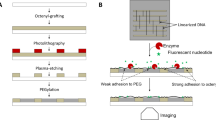
The following protocol describes the experimental steps used to prepare arrayable and label-free biological sensors that are based on the fluorescence unquenching of DNA hairpins immobilized on metal surfaces. This two-part protocol describes both the creation of gold-coated substrates and the oligonucleotide surface self-assembly process that transforms the substrates into reporters for detecting DNA. Using this procedure, one can create sensors for oligonucleotides that are highly sensitive and have demonstrated an exceptional specificity to single nucleotide polymorphisms. From start to finish, the entire procedure can be accomplished in 24–30 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Yoo, S.M. et al. Development of DNA microarray for pathogen detection. Biotech. Bioprocess Eng. 2004; 9, 93–99.
Gracey, A.Y. & Cossins, A.R. Application of microarray technology in environmental and comparative physiology. Annu. Rev. Physiol. 2003; 65, 231–259.
Guo, Q.M. DNA microarray and cancer. Curr. Opin. Oncol. 2003; 15, 36–43.
Fan, J. et al. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl. Acad. Sci. USA 2002; 99, 10611–10616.
Fang, Y., Frutos, A.G. & Lahiri, J. Ganglioside microarrays for toxin detection. Langmuir 2003; 19, 1500–1505.
Schäfering, M., Kruschina, M., Meerkamp, M., Ortigao, F. & Kambhmpati, D. Smart sensor architectures: importance of micro-array sensor surface in conducting high quality functional genomic analysis. PharmaGenomics, 2002; Sept/Oct, 36–44.
Pirrung, M.C. How to make a DNA chip. Agnew. Chem. Int. Ed. 2002; 41, 1276–1289.
Donovan, D.M. & Becker, K.G. Double round hybridization of membrane based cDNA arrays: improved background reduction and data replication. J. Neurosci. Methods 2002; 118, 59–62.
Sawiris, G.P. et al. Development of a highly specialized cDNA array for the study and diagnosis of epithelial ovarian cancer. Cancer Res. 2002; 62, 2923–2928.
Vainrub, A. & Pettitt, B.M. Sensitive quantitative nucleic acid detection using oligonucleotide microarrays. J. Am. Chem. Soc. 2003; 125, 7798–7799.
Steemers, F.J., Ferguson, J.A. & Walt, D.R. Screening unlabeled DNA targets with randomly ordered fiber-optic gene arrays. Nat. Biotechnol. 2000; 18, 91–94.
Liu, X. & Tan, W.A. A fiber-optic evanescent wave DNA biosensor based on novel molecular beacons. Anal. Chem. 1999; 71, 5054–5059.
Piestert, O. et al. A single-molecule sensitive DNA hairpin system based on intramolecular electron transfer. Nano Lett. 2003; 3, 979–982.
Fang, X., Lu, X., Schuster, S. & Tan, W. Designing a novel molecular beacon for surface-immobilized DNA hybridization studies. J. Am. Chem. Soc. 1999; 121, 2921–2922.
Du, H., Disney, M.D., Miller, B.L. & Krauss, T.D. Hybridization-based unquenching of DNA hairpins on Au surfaces: prototypical 'molecular beacon' biosensors. J. Am. Chem. Soc. 2003; 125, 4012–4013.
Du, H., Strohsahl, C.M., Camera, J., Miller, B.L. & Krauss, T.D. Sensitivity and specificity of metal surface-immobilized 'molecular beacon' biosensors. J. Am. Chem. Soc. 2005; 127, 7932–7940.
Strohsahl, C.M., Du, H., Miller, B.L. & Krauss, T.D. Towards single-spot, multianalyte molecular beacon biosensors. Talanta 2005; 67, 479–485.
Strohsahl, C.M., Miller, B.L. & Krauss, T.D. Identification of high-stringency DNA hairpin probes by partial gene folding. Biosensors Bioelectron. (in the press).
Fan, C., Plaxco, K.W. & Heeger, A.J. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl. Acad. Sci. USA 2003; 100, 9134–9137.
Lakowicz, J.R. Quenching of fluorescence. in Principles of Fluorescence Spectroscopy 3rd edn. (ed. Lakowicz, J.R.) 278–330 (Springer Science+Business Media, NY).
Storhoff, J.J., Elghanian, R., Mucic, R.C., Mirkin, C.A. & Letsinger, R.L. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J. Am. Chem. Soc. 1998; 120, 1959–1964.
He, L. et al. Colloidal Au-enhanced surface plasmon resonance for ultrasensitive detection of DNA hybridization. J. Am. Chem. Soc. 2000; 122, 9071–9077.
Dubertret, B., Calame, M. & Libchaber, A.J. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat. Biotechnol. 2001; 19, 365–370.
Tyagi, S. & Kramer, F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996; 14, 303–308.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors are the cofounders of Lighthouse Biosciences, LLC, a company based in Rochester, NY, that is commercializing this technology for medical diagnostics and homeland security applications.
Rights and permissions
About this article
Cite this article
Strohsahl, C., Miller, B. & Krauss, T. Preparation and use of metal surface-immobilized DNA hairpins for the detection of oligonucleotides. Nat Protoc 2, 2105–2110 (2007). https://doi.org/10.1038/nprot.2007.301
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.301
This article is cited by
-
Electroanalysis of single-nucleotide polymorphism by hairpin DNA architectures
Analytical and Bioanalytical Chemistry (2013)
-
Recent trends in molecular beacon design and applications
Analytical and Bioanalytical Chemistry (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.