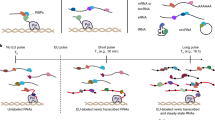
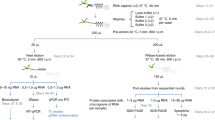
Abstract
RNA targets of multitargeted RNA-binding proteins (RBPs) can be studied by various methods including mobility shift assays, iterative in vitro selection techniques and computational approaches. These techniques, however, cannot be used to identify the cellular context within which mRNAs associate, nor can they be used to elucidate the dynamic composition of RNAs in ribonucleoprotein (RNP) complexes in response to physiological stimuli. But by combining biochemical and genomics procedures to isolate and identify RNAs associated with RNA-binding proteins, information regarding RNA–protein and RNA–RNA interactions can be examined more directly within a cellular context. Several protocols — including the yeast three-hybrid system and immunoprecipitations that use physical or chemical cross-linking — have been developed to address this issue. Cross-linking procedures in general, however, are limited by inefficiency and sequence biases. The approach outlined here, termed RNP immunoprecipitation–microarray (RIP-Chip), allows the identification of discrete subsets of RNAs associated with multi-targeted RNA-binding proteins and provides information regarding changes in the intracellular composition of mRNPs in response to physical, chemical or developmental inducements of living systems. Thus, RIP-Chip can be used to identify subsets of RNAs that have related functions and are potentially co-regulated, as well as proteins that are associated with them in RNP complexes. Using RIP-Chip, the identification and/or quantification of RNAs in RNP complexes can be accomplished within a few hours or days depending on the RNA detection method used.
*Note: In the version of the article originally published, in the last sentence of the ANTICIPATED RESULTS section, the callout should be to reference 19 instead of 18. The error has been corrected in the HTML and PDF versions of the article.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
Change history
31 August 2006
In the version of the article originally published, in the last sentence of the ANTICIPATED RESULTS section, the callout should be to reference 19 instead of 18. The error has been corrected in the HTML and PDF versions of the article.
References
Ideker, T. et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292, 929–934 (2001).
Griffin, T.J. et al. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol. Cell. Proteomics 1, 323–333 (2002).
Keene, J.D. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. USA 98, 7018–7024 (2001).
Keene, J.D. & Tenenbaum, S.A. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9, 1161–1167 (2002).
Hieronymus, H. & Silver, P.A. A systems view of mRNP biology. Genes Dev. 18, 2845–2860 (2004).
Keene, J.D. & Lager, P.J. Posttranscriptional operons and regulons coordinating gene expression. Chr. Res. 13, 327–337 (2005).
Moore, M.J. From birth to death: the complex lives of eukaryotic mRNAs. Science 309, 1514–1518 (2005).
Cilley, C.D. & Williamson, J.R. PACE Analysis of RNA-peptide interactions. In Methods in Molecular Biology (S.R. Haynes, ed.) 129–142 (Humana Press, Totowa, New Jersey, 1999)
Gao, F., Carson, C., Levine, T.D. & Keene, J.D. Selection of a subset of mRNAs from 3′UTR combinatorial libraries using neuronal RNA-binding protein, Hel-N1. Proc. Natl. Acad. Sci. USA 91, 11207–11211 (1994).
SenGupta, D.J. et al. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. USA 93, 8496–8501 (1996).
Bernstein, D.S., Buter, N., Stumpf, C. & Wickens, M. Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods 26, 123–141 (2002).
Wang, Z.F. et al. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10, 3028–3040 (1996).
Tenenbaum, S.A., Carson, C.C., Lager, P.J. & Keene, J.D. Identifying mRNA subsets in mRNP complexes using cDNA arrays. Proc. Natl. Acad. Sci. USA 97, 14085–14090 (2000).
Tenenbaum, S.A., Lager, P.J., Carson, C.C. & Keene, J.D. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods 26, 191–198 (2002).
Tenenbaum, S.A., Carson, C.C., Atasoy, U. & Keene, J.D. Genome-wide regulatory analysis combining en masse nuclear run-ons (emRUNs) and ribonomic profiling. Gene 317, 79–87 (2003).
Eystathioy, T. et al. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell 13, 1338–1351 (2002).
Kaneko, S. & Manley, J.L. The mammalian RNA polymerase II C-terminal domain interacts with RNA to suppress transcription-coupled 3′ end formation. Mol. Cell 20, 91–103 (2005).
Mili, S. & Steitz, J.A. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA 10, 1692–1694 (2004).
Penalva, L.O.F. Burdick, M.D., Lin, S.M., Sutterluety, H. & Keene, J.D. RNA-binding proteins to assess gene expression states of co-cultivated cells in response to tumor cells. Mol. Can. 3, 24–35 (2004).
Penalva, O.F., Tenenbaum, S.A. & Keene, J.D. Gene expression analysis of messenger RNP complexes. Meth. Mol. Biol. 257, 125–134 (2004).
Roy, P.J., Stuart, J.M., Lund, J. & Kim, S.K. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418, 975–979 (2002).
Niranjanakumari, S., Lasda, E., Brazas, R. & Garcia-Blanco, M.A. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26, 182–190 (2002).
Ule, J., Jensen, K., Mele, A. & Darnell, R.B. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods 37, 376–386 (2005).
Zang, Z., Edenberg, H.J. & Davis, R.L. Isolation of mRNA from specific tissues of Drosophilia by mRNA tagging. Nuc. Acids Res. 33, e148 (2005).
Kunitomo, H., Uesugi, H., Kohara, Y. & Iino, Y. Identification of ciliated sensory neuron-expressed genes in Caenorhabditis elegans using targeted pull-down of poly(A) tails. Genome Biol. 6, R17 (2005).
Ule, J. et al. CLIP identifies Nova-regulated RNA networks in the brain. Science 302, 1212–1215 (2003).
Gerber, A.P., Herschlag, D. & Brown, P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2, E79 (2004).
Gerber, A.P., Luschnig, S., Kransow, M.A., Brown, P.O. & Herschlag, D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 103, 4487–4492 (2006).
Penalva, P.O. & Keene, J.D. Biotinylated tags for recovery and characterization of ribonucleoprotein complexes. Biotechniques 37, 604–610 (2004).
Antic, D., Lu, N. & Keene, J.D. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 13, 449–461 (1999).
Zhu, J., Shendure, J., Mitra, R.D. & Church, G.M. Single molecule profiling of alternative pre-mRNA splicing. Science 301, 836–838 (2003).
Kapranov, P. et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296, 916–919 (2002).
Thomson, J.M., Parker, J., Perou, C.M. & Hammond, S.M. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 1, 47–53 (2004).
Zhang, B., Kirov, S. & Snoody, J. WebGestalt: and integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–748 (2005).
Brown, V. et al. Microarray identification of Fragile X-Mental Retardation protein (FMRP)-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107, 477–487 (2001).
Lopez de Silanes, I., Zhan, M., Lal, A., Yang, X. & Gorospe, M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA 101, 2987–2992 (2004).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
J.D.K. holds stock in Ribonomics, Inc., a company that owns patents for the RIP-Chip technology.
Rights and permissions
About this article
Cite this article
Keene, J., Komisarow, J. & Friedersdorf, M. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1, 302–307 (2006). https://doi.org/10.1038/nprot.2006.47
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.47
This article is cited by
-
RIP-PEN-seq identifies a class of kink-turn RNAs as splicing regulators
Nature Biotechnology (2024)
-
NAP-seq reveals multiple classes of structured noncoding RNAs with regulatory functions
Nature Communications (2024)
-
3D genomics and its applications in precision medicine
Cellular & Molecular Biology Letters (2023)
-
LINE-1 regulates cortical development by acting as long non-coding RNAs
Nature Communications (2023)
-
On the utility of microfluidic systems to study protein interactions: advantages, challenges, and applications
European Biophysics Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.