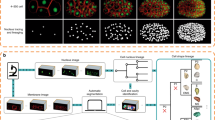
Abstract
Lineage analysis of Caenorhabditis elegans is a powerful tool for characterizing developmental phenotypes and embryonic gene-expression patterns. We present a detailed protocol for the lineaging of embryos by computational analysis of 4D images of embryos that ubiquitously express histone–GFP (green fluorescent protein) fusion proteins through the 350 cell stage followed by manual editing. We describe how to optimize imaging settings for this purpose, the use of the lineage-extraction software, StarryNite, and the lineage-editing software, AceTree. In addition, we describe a useful polymer bead mounting technique for C. elegans embryos that has several advantages compared with the standard agar pad mounting technique. The protocol requires about 1 h of user time spread over 2 days to generate the raw lineage, and an additional 2 or 4 h to edit the lineage to the 194- or 350-cell stage, respectively.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sulston, J.E. & Horvitz, H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977).
Sulston, J.E., Schierenberg, E., White, J.G. & Thomson, J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 (1983).
Schnabel, R., Hutter, H., Moerman, D. & Schnabel, H. Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Dev. Biol. 184, 234–265 (1997).
Martinelli, S.D., Brown, C.G. & Durbin, R. Gene expression and development databases for C. elegans. Semin. Cell Dev. Biol. 8, 459–467 (1997).
Bao, Z. et al. Automated cell lineage tracing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 103, 2707–2712 (2006).
Boyle, T.J., Bao, Z., Murray, J.I., Araya, C.L. & Waterston, R.H. AceTree: a tool for visual analysis of Caenorhabditis elegans embryogenesis. BMC Bioinformatics 7, 275 (2006).
Hope, I. A. (ed.) C. elegans A Practical Approach (Oxford University Press, Oxford, 1999).
Boyd, L., Guo, S., Levitan, D., Stinchcomb, D.T. & Kemphues, K.J. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegans embryos. Development 122, 3075–3084 (1996).
Acknowledgements
This research is partly funded by the US National Institutes of Health. J.I.M. is a fellow of the Jane Coffin Childs Memorial Fund for Medical Research. Z.B. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Video 1
12 cell-stage image stack. Each frame is a separate image plane starting at the top of the stack. In addition to the 12 cells at this stage, the polar body is visible as a smaller, more intense region of staining. (MOV 19033 kb)
Supplementary Video 2
100 cell-stage image stack. (MOV 3467 kb)
Supplementary Video 3
200 cell-stage image stack. (MOV 2996 kb)
Supplementary Video 4
350 cell-stage image stack. (MOV 3111 kb)
Supplementary Video 5
Full image series at plane 15. Each frame is a separate time point, 1 min apart. The images are from plane 15, near the center of the embryo. (MOV 2980 kb)
Rights and permissions
About this article
Cite this article
Murray, J., Bao, Z., Boyle, T. et al. The lineaging of fluorescently-labeled Caenorhabditis elegans embryos with StarryNite and AceTree. Nat Protoc 1, 1468–1476 (2006). https://doi.org/10.1038/nprot.2006.222
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2006.222
This article is cited by
-
Multilevel regulation of muscle-specific transcription factor hlh-1 during Caenorhabditis elegans embryogenesis
Development Genes and Evolution (2020)
-
AceTree: a major update and case study in the long term maintenance of open-source scientific software
BMC Bioinformatics (2018)
-
Comparative proteome analysis between C . briggsae embryos and larvae reveals a role of chromatin modification proteins in embryonic cell division
Scientific Reports (2017)
-
Overlapping cell population expression profiling and regulatory inference in C. elegans
BMC Genomics (2016)
-
CellProfiler Tracer: exploring and validating high-throughput, time-lapse microscopy image data
BMC Bioinformatics (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.