Abstract
The capacity to form long-lasting social memories is critical to our health and survival. cAMP signaling in the ventral hippocampal formation (VHIPP) appears to be required for social memory formation, but the phosphodiesterase (PDE) involved remains unknown. Previously, we showed that PDE11A, which degrades cAMP and cGMP, is preferentially expressed in CA1 and subiculum of the VHIPP. Here, we determine whether PDE11A is expressed in neurons where it could directly influence synaptic plasticity and whether expression is required for the consolidation and/or retrieval of social memories. In CA1, and possibly CA2, PDE11A4 is expressed throughout neuronal cell bodies, dendrites (stratum radiatum), and axons (fimbria), but not astrocytes. Unlike PDE2A, PDE9A, or PDE10A, PDE11A4 expression begins very low at postnatal day 7 (P7) and dramatically increases until P28, at which time it stabilizes to young adult levels. This expression pattern is consistent with the fact that PDE11A is required for social long-term memory (LTM) formation during adolescence and adulthood. Male and female PDE11 knockout (KO) mice show normal short-term memory (STM) for social odor recognition (SOR) and social transmission of food preference (STFP), but no LTM 24 h post training. Importantly, PDE11A KO mice show normal LTM for nonsocial odor recognition. Deletion of PDE11A may impair memory consolidation by impairing requisite protein translation in the VHIPP. Relative to WT littermates, PDE11A KO mice show reduced expression of RSK2 and lowered phosphorylation of S6 (pS6–235/236). Together, these data suggest PDE11A is selectively required for the proper consolidation of recognition and associative social memories.
Similar content being viewed by others
INTRODUCTION
What humans consider to be ‘socially acceptable behaviors’ are largely learned through social experiences. Without acceptable social behaviors, our abilities to develop meaningful friendships, attract a mate, acquire resources from society, and establish a safe/secure environment are severely compromised (Ferguson et al, 2002; Tipton et al, 2013). Thus, the capacity to form long-lasting memories of our social experiences is critical to our health and survival.
From amoebas (Gregor et al, 2010) to mammals (Bickle, 2008), cAMP /cGMP signaling appears to be a conserved molecular mechanism regulating social behavior. Oxytocin, a neuropeptide well known to regulate social behaviors (Lukas and Neumann, 2013; Stoesz et al, 2013), couples directly and indirectly to the cAMP and cGMP cascades (Cheng et al, 2009; Viero et al, 2010). Further, cAMP-response element-binding protein (CREB), exchange protein activated by cAMP (Epac), and the cyclic nucleotide-degrading phosphodiesterases (PDEs) appear to modulate various aspects of social behavior (Boess et al, 2004; Bickle, 2008; Grauer et al, 2009; Kelly et al, 2010; Hutson et al, 2011; Liebenberg et al, 2012; Srivastava et al, 2012; Yang et al, 2012). cAMP signaling in the ventral hippocampal formation (VHIPP; defined here as encompassing CA1-CA4, dentate gyrus, subiculum, and the adjacently connected amygdalohippocampal area (AHi)) may be particularly critical in the formation of social memories because knockdown of CREB function in the VHIPP impairs the formation of social memories (Brightwell et al, 2005) as does lesioning the VHIPP (Kogan et al, 2000; Tseng et al, 2009). Interestingly, both cyclic nucleotide signaling deficits and VHIPP abnormalities (referred to as anterior hippocampus in primates) have been associated with neuropsychiatric disorders in which social deficits are known to occur (c.f., (Kelly, 2015)).
Although it is known that cAMP signaling in the VHIPP is required for the formation of social memories, it is not yet known which PDE controls the compartmentalization of these signals. PDE11A, which hydrolyzes cAMP and cGMP equally well, is the only PDE to be preferentially expressed in the hippocampus (Kelly et al, 2010; Kelly et al, 2014; Kelly, 2015). We have shown that PDE11A mRNA is almost exclusively expressed in CA1, subiculum, and AHi of the VHIPP, with minimal expression in the dorsal HIPP (DHIPP) (Figure 1; (Kelly et al, 2010; Kelly et al, 2014)) and no expression elsewhere in the brain or in peripheral organs (Kelly, 2015). Importantly, PDE11A knockout (KO) mice exhibit deficits in social approach behaviors (Kelly et al, 2010; Hegde et al, 2016) and PDE11A has been genetically/functionally associated with clinical phenotypes related to changes in social function, including suicide risk (Coon et al, 2013), major depressive disorder (Wong et al, 2006; Cabanero et al, 2009; Luo et al, 2009), and lithium response (Couzin, 2008; Kelsoe, 2010; Mertens et al, 2015).
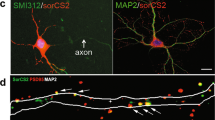
PDE11A4 is selectively expressed in neurons. Sagittal sections (~2.64 mm lateral from Bregma) were taken from PDE11A wild-type (WT) and knockout (KO) mice and stained for PDE11A4 (PD11A-112; green), nuclei (DAPI; blue), and a marker for neurons (neuron specific enolase, NSE; red), astrocytes (glial fibrillary acidic protein, GFAP; red), dendrites (MAP2; red), or axon bundles (myelin basic protein, MBP; red). (a) Staining for PDE11A4 is far stronger in hippocampi of PDE11A WT mice vs KO mice, arguing for specificity of the antibody. Further, PDE11A4 expression was far stronger in the ventral hippocampal formation (VHIPP) of PDE11A WT mice vs the dorsal hippocampal formation (DHIPP) of PDE11A WT mice, consistent with previous reports using in situ hybridization for mRNA and western blotting for protein. PDE11A4 protein can be seen in the cell body layer and throughout stratum radiatum of CA1. At the anterior edge of the CA1 field (facing left), a slightly more intense patch of staining that extends throughout stratum radiatum in the shape of a narrow triangle is reliably observed across animals. The anatomical localization of this staining suggests it may actually reflect CA2, although it is thought that CA2 is minimally present in VHIPP. Labeling in the cell body layer and stratum radiatum of ventral CA1 abruptly stops anteriorly at the border for CA3 and dorsally at the stratum lacunosum of dentate gyrus. Labeling for PDE11A4 can also be seen in the axon bundle projecting out of the hippocampus. (b) A closer view shows that PDE11A4 is expressed in a subset of neuronal cell bodies, particularly those neurons lying adjacent to the stratum radiatum. (c) In contrast, PDE11A4 does not appear to be expressed in astrocytes. (d) Consistent with its expression throughout the stratum radiatum, PDE11A4 protein expression colocalizes in some instances with the dendritic marker MAP2. (e) PDE11A4 protein expression also colocalizes with MBP. PDE11A4 protein expression in axons is consistent with the fact that faint PDE11A4 protein expression can be measured when western blotting brain regions that, themselves, do not express PDE11A mRNA but do receive projections from the hippocampus (eg, prefrontal cortex and striatum). Histogram stretch and gamma adjustments applied uniformly across PDE11A KO and WT sections for graphical clarity of images.
Previously, we showed that adult PDE11A KO mice fail to form long-term memories (LTMs) for social odor recognition (SOR), despite normal performance during training (Kelly et al, 2010). These findings suggested that PDE11A has a role in the consolidation and/or retrieval of SOR LTM. Here, we test the hypothesis that PDE11A regulates memory consolidation specifically. To do so, we compare short-term memory (STM) vs LTM and examine biochemical markers of memory consolidation in VHIPP and DHIPP of PDE11A wild-type (WT) and mutant mice. We also determine whether the SOR LTM impairment exhibited by PDE11A KO mice reflects a deficit in forming social memories specifically, which would be consistent with the fact that deletion of PDE11A alters gene expression in the oxytocin pathway (Hegde et al, 2016), or a deficit in forming recognition memories in general. To do so, we additionally test PDE11A WT and KO mice in nonsocial odor recognition (NSOR) and social transmission of food preference (STFP). Together, studies here suggest that PDE11A in the VHIPP has a specific role in the consolidation of LTMs for social experiences.
MATERIALS AND METHODS
Subjects
PDE11A knockout (KO) mice were originally developed by Deltagen (San Mateo, CA), and are maintained on a mixed C57BL/6J-C57BL/6N-129S6 background, as previously described (Kelly et al, 2010). The deletion targets the catalytic domain and, therefore, targets all isoforms of PDE11A (not only PDE11A4, which is the only isoform expressed in brain). PDE11A mice were bred onsite in heterozygous (HT) × HT matings, with same-sex WT, HT, and KO littermates weaned together. There is essentially no concern of litter effects driving the effects described herein as any given litter typically contributes n=1–2 mice/genotype to a cohort (because of the use of sex-matched littermates) and, at most, parents contribute 2 litters to a cohort (ie, a total of 2–4 mice/genotype). Animals are housed on a 12:12 light:dark cycle and allowed ad lib access to food and water with select exceptions (see below). Approximately equal numbers of male and female offspring were tested between P28 and P42 for adolescent studies and 2 and 12 months of age for adult studies. Multiple cohorts of mice were tested; some in multiple assays with at least 1 week between tests (see Supplementary Table). In all experiments, PDE11A KO mice were compared with sex-matched littermates. See figure legends for experimental n’s. In all experiments, genotypes were counterbalanced across technical variables (eg, bead position, spice combination and placement, position on gel, and so on). Experiments were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (Pub 85–23, revised 1996) and were fully approved by the Institutional Animal Care and Use Committee of the University of South Carolina.
Biochemistry and Molecular Biology
Molecular biology experiments were conducted as previously described (Kelly et al, 2010; Kelly et al, 2014; Pathak et al, 2015). Animals were killed by cervical dislocation for studies in Figure 1 and Figure 4 and by in vivo microwave irradiation (Muromachi In Vivo Microwave, 4 kW for 1.1 s) for studies in Figure 5. For western blots, freshly frozen tissue was sonicated in ice-cold lysis buffer (20 mM Tris-HCl, pH 7.5; 2 mM MgCl2; Thermo Pierce Scientific phosphatase and protease inhibitor tablet) (Figure 1), and the microwaved tissue was sonicated in boiling lysis buffer (50 mM NaF/1% SDS). Equal amounts of total protein were separated by gel electrophoresis using Invitrogen 4–12% Bis-Tris gels (Life Technologies). Following transfer, nitrocellulose membranes were probed with antibodies for PDE11A (Fabgennix PD11–112 Lot 173, 1:500 (Kelly et al, 2010)), S6 (Cell Signaling 2217S, 1:500), pS6–235/236 (Cell Signaling 4856S, 1:500), pS6–240/244 (Cell Signaling 4838S, 1:500), ribosomal S6 kinase 2 (RSK2; Phosphosolutions 1850-rsk2, 1:10 000); actin as a loading control (Sigma #A2066; 1:10 000) and an HRP-tagged secondary antibody recognizing rabbit IgG (Jackson Immunoresearch #111-035-144; 1:10 000). Multiple film exposures were taken to ensure data were collected within the linear range. Western blots were quantified using Image J (NIH).
Immunofluorescence
Immunofluorescence was conducted based on the method of Wood et al (2009), with modification. Animals were killed by cervical dislocation. Brains were harvested fresh and frozen at −80 °C until processing. Two PDE11A WT and two KO brains were blocked in embedding matrix, cryosectioned in the sagittal plane at 20 μm, and thaw-mounted onto the same slide. Slides were air-dried and then stored at −80 °C until processing. Slides were fixed for 20 min in 4% paraformaldehyde (in water or 1 × phosphate-buffered saline depending on antibody) and then washed 3 × in 1 × phosphate-buffered saline. Slides were blocked with 1 × phosphate-buffered saline/0.4% BSA/0.3% Triton-X 100 (a.k.a. PBT) 3 × 10 min and then either PBT/10% BlokHen (Aves BH-1001—Tigard, OR) or PBT/5% milk for 1 h. Slides were probed overnight at 4 °C with antibodies against PDE11A (1:100 of FabGennix PD11–112- Frisco, TX), PDE11A4 (1:10 000 of Aves PDE11A#1), NSE (1:500 of Aves), MBP (1:100 of Aves), GFAP (1:500 of Aves), or MAP2 (1:2000 of Neuromics- CH22103—Minneapolis, MN). Primary antibodies were diluted in PBT, except for PDE11A#1 that was prepared in PBT/5% milk, with 0.02% NaN3. The next day, sections were washed 4 × 10 min in PBT. To detect PDE11A4 expression, slides were probed with an Alexafluor 488-tagged secondary antibody for 90 min at room temperature (1:400 of Jackson Immunoresearch 711-545-152 or 1:1000 of Jackson Immunoresearch 703-545-155- West Grove, PA). Slides were then washed in PBT 3 × 10 min. To detect the cell markers, slides were probed with a Cy3-tagged secondary antibody for 90 min at room temperature (1:1000 of Jackson Immunoresearch 703-165-155). Slides were again washed in PBT 3 × 10 min and dipped 10 times in 1 × phosphate-buffered saline to remove any detergent. Sections were mounted using a coverslip and fluoromount G (Fisher 0100-20—Pittsburgh, PA). Images were taken using a Nikon Eclipse 80 i bright-field microscope, MBF Bioscience CX9000 camera, and Neurolucida software (MBF Bioscience, Willston, VT).
Odor Recognition
The odor recognition test is based on the fact that mice explore novel odors more so than familiar odors (ie, odors they remember from the past) (Kelly et al, 2010). Mice lived with 1-inch wooden beads (home-cage odor) for 1 week and then were habituated (1 × 3-min trial), trained (2 × 3-min trials, 5-min inter-trial interval) and tested (2-min trial) >1 h after being transferred to individual clean home cages (see Figure 2a for diagram of paradigm). During habituation, mice were given 3 of their own home-cage beads. During training, mice were given 2 home-cage beads and 1 scented bead (social odors: same-sex C57BL/6J, BALB/cJ, or 129S6/SvEv; nonsocial odors: turmeric, basil, ginger). Different mouse strains were used as social odor donors, as opposed to different individual mice all of the same strain, because we find inbred mice do not differentiate social odors between mice of the same inbred strain (see Supplementary Figure 1). During testing, mice were given 1 home-cage bead and 2 scented beads (1 familiar from training and 1 novel). The amount of time investigating each bead was recorded by an experimenter blind to genotype and the novelty of the beads. Testing data are then expressed as the % time spent exploring each bead (ie, home cage or familiar or novel time/total exploration time). Odor recognition memory for the trained scent is operationally defined as subjects spending significantly more time exploring the novel vs familiar scent.
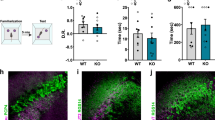
Adult PDE11A knockout (KO) mice show deficits in long-term memory (LTM) for social odor recognition (SOR) but not nonsocial odor recognition (NSOR). (a) A diagram of the SOR and NSOR paradigms are shown, where ‘memory’ is operationally defined as spending significantly more time investigating the novel vs the familiar odor. (b) PDE11A wild-type (WT) and knockout (KO) mice exhibit significant short-term memory (STM) for SOR 1 h after training. (c) In contrast to their WT littermates, PDE11A KO mice exhibit absolutely no LTM for SOR 24 h after training. (d) This LTM deficit appears to be selective for social odors because PDE11A KO mice demonstrate robust NSOR memory 24 h after training, and this memory is equally as strong as that shown by their WT littermates. (b), n=32–38/genotype; (c), n=18–21/genotype; (d), n=29–31/genotype. Post hoc, *vs familiar and home-cage odors, P<0.001; vs #WT within odor, P<0.03–0.001.
Social Transmission of Food Preference (STFP)
On the basis of the study by Wrenn et al (2003), food was presented to mice using 2-oz glass jars (Fisher Scientific 02-911-773) that had six equally spaced 1-cm holes drilled into the lid. The jar was filled with bedding in order to press a small weigh pan (Fisher Scientific 08-732-116) containing powdered food (standard chow pulverized in a blender) against the underside of the jar lid. For training schematic, see Figure 3a. On day1, food access was restricted to 1 h. On day2, mice were individually habituated to the powdered chow for 1 h in a clean home cage. On day3, the demonstrators were allowed to eat powdered food laced with one of two household spices used/experiment for 1 h (2% basil vs 2% ginger, 4% marjoram vs 1% cumin, or 2% basil vs 1.5% thyme, 1% cardamom vs 1% mint). Note, we were unable to use cinnamon vs cocoa in this assay, as it is typically reported, because we found females to invariably eat the cinnamon-laced food. The demonstrator was then returned to its home cage and observers were allowed to investigate the demonstrator for 15 min, after which time the demonstrator was removed. An experimenter verified that all observers equally interacted with the demonstrator. To test memory for STFP, observers were individually transferred to a clean cage and given 1 h to eat from two glass jars set at either end of the cage—one containing food scented with the same spice eaten by their demonstrator (ie, trained food) and the other containing a food scented with the spice assigned to another cage’s demonstrator (ie, novel food). The amount of food eaten was weighed and the time spent eating each food was recorded for 1 min every 10 min by an experimenter blind to genotype and food type. Mice maintained ad lib access to water.
Adult PDE11A knockout (KO) mice show normal short-term memory (STM) for social transmission of food preference (STFP) but no long-term memory (LTM). (a) A diagram of the STFP assay is shown, where memory is operationally defined as mice demonstrating a significant preference for the trained vs novel food. Both PDE11A wild-type (WT) and KO mice show normal STM for STFP 10 min after training, whether measuring preference in terms of (b) the amount of each food eaten or (c) the time spent eating the trained vs novel food. In contrast, PDE11A KO mice show no LTM for STFP 24 h after training whether measuring preference in terms of (d) the amount of each food or (e) the time spent eating the trained vs novel. (b and c), n=28–34/genotype; (d and e), n=24/genotype. Post hoc, *trained vs novel, P<0.001; #WT vs KO within novel or trained, P⩽0.002–0.001.
Data Analyses
Behavioral data were analyzed for effect of genotype and sex by ANOVA or repeated-measure ANOVA using Sigmaplot 11.0 or Statistica. Biochemical data were analyzed by Student t-test for effect of genotype using Sigmaplot 11.0, as the effect of sex was negated in western blots via normalization (see below). The effect of genotype did not differ as a function of sex in any behavioral experiment (ie, genotype × sex interactions were not significant); therefore, data were graphed collapsed across sexes for sake of clarity. As previously described (Kelly et al, 2010; Kelly et al, 2014), western blot data spanned multiple blots (each blot contained all males or all females) and so were normalized to the WT mean to account for any difference in transfer efficiency, antibody binding, film exposure, and so on. Statistical outliers>2 standard deviations from the mean were removed from analyses, as previously described (Kelly et al, 2008; Kelly et al, 2010; Kelly et al, 2014) (Figure 4e, 4/61 mice; Figure 5a, 4/86 data points; Figure 5b, 1/86 data points; Figure 5c; 1/84 data points; Figure 5d, 3/86 data points; Figure 5e, 2/84 data points; Figure 5f, 3/71 data points). Significance was determined as P<0.05.
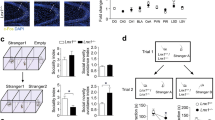
Adolescent PDE11A knockout (KO) mice demonstrate impairments in the consolidation of long-term memories (LTMs) for social experiences. (a) As measured in C57BL/6J and PDE11A wild-type male and female mice, PDE11A4 mRNA expression significantly increases in ventral CA1 and subiculum (vCA1, vSub) between postnatal day 7 (P7) and P21 (n=4–6/age). The specificity of the probe was confirmed using sections from PDE11A knockout (KO) mice. Whereas, PDE11A4 mRNA levels then stabilize (vCA1), or subsequently decrease (vSub), between P21 and P28, (b) PDE11A4 protein expression in the hippocampus significantly increases every week from P7 to P28 (n=4–6/age). No further increase in expression is observed between P28 and young adulthood (n=11–14/age). The specificity of the antibody was verified using hippocampal tissue from PDE11A KO mice. Consistent with the fact that adolescent PDE11A4 protein expression is equivalent to levels observed in young adults, adolescent PDE11A KO mice exhibit similar phenotypes to adult PDE11A KO mice. (c) Adolescent PDE11A KO mice show normal short-term memory (STM) 1 h after social odor recognition (SOR) training but (d) no LTM 24 h after SOR training (n=22–30/genotype). (e) Similarly, adolescent PDE11A KO mice show no LTM for social transmission of food preference (n=19–25/genotype). Post hoc, *vs P7, familiar odor, or novel food, P<0.005–0.001; #vs P14 or WT within odor/food, P<0.02–0.001. @vs P21, P<0.001. Brightness and contrast adjusted for graphical clarity.
Deleting PDE11A4 reduces ribosomal protein S6 phosphorylation, a biomarker of protein translation. (a) Relative to PDE11A WT littermates, PDE11A KO mice show no significant differences in expression of the ribosomal protein S6 (migrating ~32 kDa). (b) In contrast, PDE11A KO mice show a significant reduction in phosphorylation of S6 at serines 235/236 (p-235) in the ventral hippocampal formation (VHIPP) but not the dorsal hippocampal formation (DHIPP) whether pS6–235/236 is normalized by actin or (c) the total expression of S6. (d and e) The loss of pS6–235/236 appears to be selective as no difference was observed between PDE11A WT and KO mice in terms of pS6–240/244 (p-240). (f) PDE11A KO mice also show significantly reduced expression of an enzyme that phosphorylates S6 at serines 235/236 but not 240/244, namely ribosomal S6 kinase 2 (RSK2; migrating ~90 kDa). Relative to WT littermates, PDE11A KO mice show significantly reduced RSK2 expression in VHIPP but not DHIPP. (g) Representative western blots. VHIPP: n=18–19/genotype; DHIPP: n=24–26/genotype. *vs WT, P<0.05–0.001. Brightness and contrast adjusted for graphical clarity of blot images.
RESULTS
PDE11A4 is Selectively Expressed in Neurons, with Expression Throughout the Dendrites, Cell Bodies, and Axons
Previously, using autoradiographic in situ hybridization, we showed that PDE11A4 mRNA is preferentially expressed in CA1, subiculum, and the AHi in brain (Kelly et al, 2010; Kelly et al, 2014; Kelly, 2015). In order to better understand how PDE11A4 may be regulating synaptic plasticity, we used immunofluorescence to identify the cell type and subcellular localization of PDE11A4. PDE11A4 immunofluorescence in PDE11A WT brains confirms PDE11A4 protein is enriched in the VHIPP (Figure 1a). PDE11A4 appears to be confined to a subset of neurons in CA1 (and, possibly, CA2; Figure 1b) and AHi (Supplementary Figure S2B), but expressed in most neurons in subiculum (Supplementary Figure S2D). PDE11A4 is not found in astrocytes (Figure 1c, Supplementary Figure S1C and S1E). In CA1, PDE11A4 is expressed throughout the dendrites, cell bodies, and axons (Figure 1b, d, and e). A similar staining pattern is observed using immunohistochemistry (Supplementary Figure S3) as well as a PDE11A4 isoform-specific antibody in immunofluorescence (Supplementary Figure S4). In AHi and subiculum, however, PDE11A4 expression is restricted to cell bodies and axons (Supplementary Figure S2). Expression of PDE11A4 in axons is consistent with the fact that extremely low levels of PDE11A4 protein expression are detected when western blotting brain regions that, themselves, do not express PDE11A4 mRNA but do receive axonal projections from VHIPP (eg, prefrontal cortex and striatum (Kelly, 2015)). Western blotting shows hippocampal PDE11A4 protein expression does not differ between males and females as measured in adult whole hippocampus (n=10–11/sex; WT-F 1.00±0.03A.U; WT-M 0.996±0.02A.U), VHIPP (n=12/sex; WT-F 1.00±0.11A.U; WT-M 0.915±0.12A.U) or DHIPP (n=6/sex; WT-F 1.00±0.14A.U; WT-M 0.968±0.12A.U). Further, there does not appear to be a compensatory upregulation of other closely related PDEs in response to PDE11A deletion, as neither PDE2A, PDE10A, nor PDE9A protein expression differ between adult PDE11A WT vs KO littermates in the hippocampus (Supplementary Figure S5).
Adult PDE11A KO Mice Show Normal STM but No LTM for SOR
To determine whether PDE11A4 has a role in the consolidation and/or retrieval of SOR LTM, we compared STM vs LTM for SOR in PDE11A WT and KO mice. STM was tested in two separate cohorts of PDE11A mice with identical results, so combined analyses are shown in Figure 2. Relative to PDE11A sex-matched littermates, PDE11A KO mice perform normally across SOR training trials (Supplementary Figure S6A) and the STM test (effect of bead: F(2,132)=225.3, P<0.001; Figure 2a). Despite demonstrating robust STM, PDE11A KO mice show no LTM 24 h following SOR training (effect of genotype × bead: F(2,10)=10.91, P<0.001; Figure 2b), a time point at which PDE11A WT littermates demonstrate robust LTM for SOR. These data confirm our previous report showing that PDE11A KO mice showed no LTM for SOR 24 h after training (Kelly et al, 2010). There were no significant interactions between sex and genotype in any phase of testing, consistent with the fact that PDE11A4 protein expression is equivalent in males and females. The fact that STM remains intact but LTM is impaired in the PDE11A KO mice suggests that PDE11A does not have a role in SOR memory retrieval per se, but rather memory consolidation.
Adult PDE11A KO Mice Show Normal LTM for NSOR
To determine whether the SOR LTM impairment exhibited by PDE11A KO mice reflects a deficit in forming social memories specifically or a deficit in forming recognition memories in general, we tested PDE11A KO mice in NSOR. Identical results were obtained in two separate cohorts of mice, the combined analyses of which are shown in Figure 2c. Relative to PDE11A sex-matched littermates, male and female PDE11A KO mice perform normally across NSOR training trials (Supplementary Figure 6B) and the LTM test (effect of bead: F(2,112)=73.62, P<0.001; Figure 2c). The fact that LTM remains intact for NSOR but not SOR suggests that PDE11A has a specific role in the consolidation of memories involving social stimuli.
Adult PDE11A KO Mice Show Normal STM but No LTM for STFP
To determine whether PDE11A also has a role in associative memories involving social information, we tested PDE11A WT and KO mice in STFP. In STFP, the subject makes an association between a nonsocial odor (ie, a household spice) and pheromones in the cage mate’s breath (Galef et al, 1988; Munger et al, 2010), the memory of which lets the subject know a food is safe to eat. Three separate cohorts of mice were tested in STM and two separate cohorts were tested in LTM for STFP. Identical results were obtained across the multiple cohorts; therefore, data were combined for the final analyses shown in Figure 3. PDE11A KO and WT mice eat the same amount of food during the STM (WT 0.84 ±0.06 g; KO 0.91 ±0.05 g) or LTM tests (WT 0.97 ±0.09 g, KO 0.84 ±0.06 g). Relative to PDE11A sex-matched littermates, male and female PDE11A KO mice perform normally in a STM test (effect of food on preference by weight: F(1,58)=44.00, P<0.001; effect of food on preference by time: F(1,58)=67.82, P<0.001; Figure 3b and c), suggesting the PDE11A KO mice are capable of initially learning and retrieving the associative memory. Despite demonstrating robust STM, however, male and female PDE11A KO mice show no LTM 24 h following STFP training (effect of food × genotype on preference by weight: F(1,44)=12.79, P<0.001; effect of food × genotype on preference by time:: F(1,44)=18.13, P<0.001; Figure 3d and e), a time point at which PDE11A WT littermates demonstrate robust LTM for STFP. Together, the SOR and STFP data suggest that PDE11A4 is required for proper memory consolidation of both recognition and associative memories involving social information.
PDE11A4 is also Required for Social Memory Consolidation during Adolescence
Social approach behaviors become reliably measurable in the mouse around P19 (Laviola et al, 1994; Fairless et al, 2012). Therefore, we conducted autoradiographic in situ hybridization and western blot analyses to determine whether the developmental time course of PDE11A4 mRNA and protein expression parallels the emergence of reliably measured social approach behaviors in the mouse. Indeed, PDE11A4 mRNA (effect of age × region: F(9,51)=10.43, P<0.001) and protein expression in the hippocampus begins very low on P7, but dramatically increases with each passing week (effect of age: F(3,19)=67.59, P<0.001; Figure 4a and b). By P21, PDE11A4 protein expression increases 2.5-fold over P7 levels, reaching ~70% of expression measured in young adult mice. This dramatic rise in ventral hippocampal expression during the early postnatal days appears to be somewhat unique to PDE11A4. PDE2A shows only minimal increases in mRNA expression in dorsal CA1 (dCA1), dCA3, and the dentate gyrus subfields after P7 (effect of age × region: F(21,118)=3.33, P<0.001; Supplementary Figure S7A). PDE9A shows significant decreases in mRNA expression in dCA1, vCA3, and dCA3 between P7 and P14 (effect of age × region: F(21,119)=4.91, P<0.001; Supplementary Figure S7B). Finally, PDE10A shows a trend toward an increase in mRNA expression across hippocampal subfields between P7 and P21 (effect of age: F(3,119)=2.90, P=0.065; Supplementary Figure S7C). By P28, PDE11A4 expression stabilizes to young adult levels, as verified in two separate cohorts of mice (Figure 4b).
Given that PDE11A4 expression stabilizes to young adulthood expression levels around P28 (Figure 4b), we hypothesized that PDE11A4 would be required for the consolidation of social memories during adolescence (defined here as P28–P42). Just as is observed in adults, adolescent PDE11A KO mice show normal STM for SOR relative to sex-matched WT littermates (effect of bead: F(2,146)=240.28, P<0.001; Figure 4c), but no LTM for SOR (effect of bead × genotype: F(4,148)=3.62, P=0.008; Figure 4d). Similarly, adolescent PDE11A KO mice show no LTM for STFP 24 h following training, a time point at which their sex-matched WT littermates show a robust LTM for STFP (effect of food × genotype: F(2,53)=6.37, P=0.003; Figure 4e). As is observed in the adult mice, there is no effect of genotype on the total amount of food eaten during the STFP LTM test (WT, 0.96±0.07 g; HT 0.86±0.06 g, KO 0.88±0.04 g). These data suggest that PDE11A4 is required for proper social memory consolidation during both adolescence and adulthood.
Deletion of PDE11A Decreases pS6–235/236 in the VHIPP
Given that PDE11A KO mice exhibit a specific impairment in memory consolidation and memory consolidation requires de novo macromolecular synthesis, we determined whether deletion of PDE11A may impair phosphorylation of S6—a biomarker of protein synthesis. There were no significant differences between PDE11A KO mice vs WT littermates in terms of total expression of the ribosomal protein S6 in either VHIPP or DHIPP (Figure 5a). In contrast, there was a significant reduction in phosphorylation of S6 at serines 235/236 in the VHIPP, but not DHIPP, of PDE11A KO mice vs WT littermates (pS6–235/236/actin: t(14)=3.62, P=0.003; pS6–235/236/S6: t(17)=4.04, P<0.001; Figure 5b and c). The effect of PDE11A deletion on pS6–235/236 appears to be site-specific because phosphorylation of S6 at serines 240/244 did not change in either VHIPP or DHIPP (Figure 5d and e). This specific loss of phosphorylation of residue 235/236 is consistent with PDE11A KO mice also showing reduced VHIPP expression of RSK2 (t(16)=2.15, P=0.047), an enzyme that phosphorylates S6–235/236 but not S6–240/244 (Figure 5f) (Gangarossa and Valjent, 2012).
DISCUSSION
Here, we show that PDE11A4 is required for the proper consolidation of both recognition and associative memories for social experiences. PDE11A KO mice demonstrate intact STM for SOR and STFP but no LTM 24 h after training. The fact that STM remains intact but LTM is impaired suggests that PDE11A KO mice retain their ability to learn social information and retrieve a social memory, but differ in their ability to transition that social memory from short-term to long-term status. Whereas STM largely relies on proteins that already exist at the synapse, transitioning a memory to long-term status requires de novo transcription and protein translation (c.f., (Owen and Brenner, 2012; Rosenberg et al, 2014)). In the VHIPP, PDE11A KO mice show a significant reduction in pS6–235/236, a commonly used biomarker of protein translation (Figure 5b and c). It is important to note that PDE11A4, the isoform that is expressed in brain, is ~95% homologous across mouse, rat, and human (Yuasa et al, 2001a; Yuasa et al, 2001b). This high degree of homology argues that our results obtained in rodent models will translate across species. The fact that PDE11A is critical for the formation of social memories is consistent with the fact that PDE11A has been genetically associated with clinical phenotypes related to changes in social function, including suicide risk (Coon et al, 2013), major depressive disorder (Wong et al, 2006; Cabanero et al, 2009; Luo et al, 2009), and lithium responsivity (Couzin, 2008; Kelsoe, 2010).
PDE11A appears to be required for the consolidation of social memories but not nonsocial memories. Here, we show that LTM for NSOR remains intact in the PDE11A KO mice (Figure 2d), and we showed previously that PDE11A KO mice exhibit normal LTM for contextual fear conditioning (Kelly et al, 2010). The fact that PDE11A4 is required for the consolidation of social memories is consistent with the fact that PDE11A4 is enriched in the VHIPP and the VHIPP is known to be a critical neural circuit underlying the formation of social memories (Kogan et al, 2000; Brightwell et al, 2005; Tseng et al, 2009). It is also consistent with the fact that deletion of PDE11A significantly alters gene expression in the oxytocin pathway (Hegde et al, 2016). Indeed, a selective amnesia for social memories has similarly been reported in oxytocin KO mice (Ferguson et al, 2000). It will be of interest to future studies to explicitly test whether memory deficits observed in PDE11A mutant mice are directly related to impairments in oxytocin signaling.
Although adolescent PDE11A KO mice exhibit adult-like memory impairments in SOR and STFP, they show normal social approach behaviors in social contexts where adult PDE11A mutant mice show reduced social approach (Hegde et al, 2016). This raises the interesting possibility that social interaction deficits that emerge in adult PDE11A mutant mice are actually driven by their fundamental inability to form long-term memories for what is and what is not a socially acceptable behavior earlier in life. Indeed, an inability to learn appropriate social skills early in development has been linked to lower quality friendships later in adolescence among humans with intellectual disabilities (Tipton et al, 2013).
Our work argues that PDE11A4 activators may prove therapeutic in the context of social cognitive deficits. The extensive work of others suggests that PDE11A activators may also hold promise for treating various types of tumors (c.f., (Kelly, 2015)). In a number of PDE families, cyclic nucleotide binding of the N-terminal regulatory GAF domain can alter catalytic activity of the enzyme (Francis et al, 2011). Indeed, there is proof of principle for therapeutically targeting GAF domains (Schultz et al, 2011), and it has proven possible to stimulate PDE11A4 via its GAF-A domain with Rp-8-pCPT-cGMPS (Jager et al, 2012). Unfortunately, the use of this particular compound as a tool PDE11A activator is severeley limited by off-target activities (namely inhibition of PKG and, likely, many other cGMP-binding molecules). That said, the ability of Rp-8-pCPT-PET-cGMPs to allosterically stimulate PDE11A function does provide an exciting proof of concept for how we might target PDE11A in order to increase catalytic activity. PDE11A4 GAF domains are less than 50% homologous to those in other PDE families (Yuasa et al, 2000), and PDE11A1–3 do not contain a full GAF-A domain (Yuasa et al, 2000). This suggests that a compound targeting the PDE11A4 GAF-A domain will not only be selective relative to other PDE families, but will also be selective for activating PDE11A4 but not PDE11A3, A2 or A1. Such selectivity would reduce side effect potential that could be associated with targeting PDE11A signaling outside the brain.
Interestingly, expression of PDE11A4 in the HIPP is minimal before P19 in the mouse (Figure 4a and b), which is the approximate age when social approach behavior can first be reliably measured in a mouse (Laviola et al, 1994; Fairless et al, 2012). We show a fivefold increase in hippocampal PDE11A4 protein expression between P7 and P28 (Figure 4b). PDE11A4 protein expression levels then stabilize for a period of time, with equivalent levels of expression observed in P28 vs young adult mice (Figure 4b). This dramatic increase in PDE11A4 protein expression early in life is reminiscent of that observed later in life in the aged rodent brain (Kelly et al, 2014). This suggests that PDE11A4 function in brain evolves across the lifespan; therefore, age should be considered when interpreting the effects of PDE11A gain- or loss-of-function.
PDE11A4 is clearly not expressed in astrocytes (Figure 1c,Supplementary Figure S2C and S2E) but, rather, is expressed in a subset of neurons in CA1 (Figure 1b) and AHi (Supplementary Figure S2B) but most neurons of ventral subiculum (Supplementary Figure S2D). Within that subset of CA1—and, possibly, CA2—neurons, PDE11A4 is expressed throughout the cell bodies, dendrites, and axons (Figure 1b, d, and e). Thus, PDE11A4 is positioned to shape signals both coming into and leaving CA1. The VHIPP, particularly ventral subiculum, is well known to be required for the consolidation of social memories (Kogan et al, 2000; Brightwell et al, 2005; Tseng et al, 2009). A role for AHi and CA2 are less well understood. Clearly, dorsal CA2 is required for social STMs (eg, Hitti and Siegelbaum (2014)); however, the role of ventral CA2 has yet to be interrogated. It will be important for future studies to determine whether the PDE11A-expressing subpopulation of CA1 and AHi neurons noted herein reflects a specific biochemically defined subpopulation (eg, inhibitory vs excitatory) and/or a specific anatomically defined subpopulation of neurons (eg, neurons projecting to a specific target region).
Cyclic nucleotides are not found uniformly throughout the cell, but instead are restricted to discrete pockets (a.k.a. microdomains), allowing a single cell to respond discretely to multiple incoming extracellular signals. PDEs have an important role in maintaining the integrity of these cyclic nucleotide microdomains. PDE11A degrades cAMP and cGMP, and we previously measured a significant decrease in cAMP-PDE activity in VHIPP of PDE11A KO mice vs WT littermates (Kelly et al, 2010). As such, we expect that PDE11A KO mice exhibit increases in these cyclic nucleotides within specific microdomains of neurons that would normally express PDE11A4; however, we have struggled to measure changes in cAMP/cGMP at the level of the whole VHIPP (Kelly et al, 2010). This may be related to the sparse nature of PDE11A4 expression in the VHIPP (Figure 1,Supplementary Figure S1), or it may be related to the fact that focal increases in cAMP/cGMP can lead to distal reductions in cyclic nucleotide signaling due to compensation by other parts of the machinery (eg, other cyclases or PDEs), particularly in the hippocampus (Kelly et al, 2007; Kelly et al, 2008; Kelly et al, 2009). Indeed, we have previously reported evidence of increased CREB phosphorylation (Pathak et al, 2016) yet decreased Glur1 phosphorylation in VHIPP of PDE11A KO mice vs WT littermates (Kelly et al, 2010), which would be consistent with there being both localized increases and localized decreases in PKA/PKG signaling in the PDE11A KOs. Given this fact, coupled with the restricted nature of PDE11A4 expression within the VHIPP, we are doubtful that a global modulation of cAMP levels throughout the entire VHIPP would rescue the memory deficits observed herein.
In conclusion, we show here that PDE11A is selectively expressed in neurons and is required for the proper consolidation of social but not nonsocial memories. Such a selective behavioral role for PDE11A is consistent with the fact that PDE11A4 is enriched in the VHIPP. Although PDE11A4 expression is largely restricted to the ventral hippocampus, and the biochemical changes noted herein occur in ventral but not dorsal hippocampus, it is not yet clear whether PDE11A4 in VHIPP and/or DHIPP is required for the consolidation of social memories. It will be important to future studies to target each subregion individually to discern where within the hippocampus PDE11A4 modulates these memory processes. Further, we cannot eliminate the possibility that other effects of PDE11A deletion have been masked by a compensatory upregulation of other signaling molecules, particularly given the constitutive nature of our KO. Thus, future studies examining more acute modulation of PDE11A4 will be of interest. Although more work remains, our findings lend further support for considering PDE11A, a highly druggable enzyme, as a potential novel therapeutic target (Kelly et al, 2010; Kelly, 2015; Hegde et al, 2016).
FUNDING AND DISCLOSURE
MPK received consulting fees from ASUBIO, Inc. and Deallus for projects unrelated to the current manuscript. SH, WRC, BAI, JK, NSP and ATS have no financial conflicts to disclose. Research supported by a Research Starter Grant in Pharmacology & Toxicology from the PhRMA Foundation, an ASPIRE award from the Office of the Vice President for Research from the University of South Carolina, a Research Development Fund Award from the University of South Carolina School of Medicine, 1R01MH101130 from NIMH, and a NARSAD Young Investigator Award from the Brain & Behavior Research Foundation (all awards to MPK).
References
Bickle J (2008). The molecules of social recognition memory: implications for social cognition, extended mind, and neuroethics. Conscious Cogn 17: 468–474.
Boess FG, Hendrix M, van der Staay FJ, Erb C, Schreiber R, van Staveren W et al (2004). Inhibition of phosphodiesterase 2 increases neuronal cGMP, synaptic plasticity and memory performance. Neuropharmacology 47: 1081–1092.
Brightwell JJ, Smith CA, Countryman RA, Neve RL, Colombo PJ (2005). Hippocampal overexpression of mutant creb blocks long-term, but not short-term memory for a socially transmitted food preference. Learn Mem 12: 12–17.
Cabanero M, Laje G, Detera-Wadleigh S, McMahon FJ (2009). Association study of phosphodiesterase genes in the Sequenced Treatment Alternatives to Relieve Depression sample. Pharmacogenet Genomics 19: 235–238.
Cheng CY, Chu JY, Chow BK (2009). Vasopressin-independent mechanisms in controlling water homeostasis. J Mol Endocrinol 43: 81–92.
Coon H, Darlington T, Pimentel R, Smith KR, Huff CD, Hu H et al (2013). Genetic risk factors in two Utah pedigrees at high risk for suicide. Transl Psychiatry 3: e325.
Couzin J (2008). Science and commerce. Gene tests for psychiatric risk polarize researchers. Science 319: 274–277.
Fairless AH, Dow HC, Kreibich AS, Torre M, Kuruvilla M, Gordon E et al (2012). Sociability and brain development in BALB/cJ and C57BL/6J mice. Behav Brain Res 228: 299–310.
Ferguson JN, Young LJ, Hearn EF, Matzuk MM, Insel TR, Winslow JT (2000). Social amnesia in mice lacking the oxytocin gene. Nat Genet 25: 284–288.
Ferguson JN, Young LJ, Insel TR (2002). The neuroendocrine basis of social recognition. Front Neuroendocrinol 23: 200–224.
Francis SH, Blount MA, Corbin JD (2011). Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions. Physiol Rev 91: 651–690.
Galef BG Jr ., Mason JR, Preti G, Bean NJ (1988). Carbon disulfide: a semiochemical mediating socially-induced diet choice in rats. Physiol Behav 42: 119–124.
Gangarossa G, Valjent E (2012). Regulation of the ERK pathway in the dentate gyrus by in vivo dopamine D1 receptor stimulation requires glutamatergic transmission. Neuropharmacology 63: 1107–1117.
Grauer SM, Pulito VL, Navarra RL, Kelly MP, Kelley C, Graf R et al (2009). Phosphodiesterase 10A inhibitor activity in preclinical models of the positive, cognitive, and negative symptoms of schizophrenia. J Pharmacol Exp Ther 331: 574–590.
Gregor T, Fujimoto K, Masaki N, Sawai S (2010). The onset of collective behavior in social amoebae. Science 328: 1021–1025.
Hegde S, Oliver D, Poupore N, Shtutman M, Kelly MP (2016). Pde11a is required for intact social behaviors and is a key mechanism by which social experience sculpts the brain. Neuropharmacology (under review).
Hitti FL, Siegelbaum SA (2014). The hippocampal CA2 region is essential for social memory. Nature 508: 88–92.
Hutson PH, Finger EN, Magliaro BC, Smith SM, Converso A, Sanderson PE et al (2011). The selective phosphodiesterase 9 (PDE9) inhibitor PF-04447943 (6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyran-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one) enhances synaptic plasticity and cognitive function in rodents. Neuropharmacology 61: 665–676.
Jager R, Russwurm C, Schwede F, Genieser HG, Koesling D, Russwurm M (2012). Activation of PDE10 and PDE11 phosphodiesterases. J Biol Chem 287: 1210–1219.
Kelly MP (2015). Does phosphodiesterase 11A (PDE11A) hold promise as a future therapeutic target? Curr Pharm Des 21: 389–416.
Kelly MP, Adamowicz W, Bove S, Hartman AJ, Mariga A, Pathak G et al (2014). Select 3',5'-cyclic nucleotide phosphodiesterases exhibit altered expression in the aged rodent brain. Cell Signal 26: 383–397.
Kelly MP, Cheung YF, Favilla C, Siegel SJ, Kanes SJ, Houslay MD et al (2008). Constitutive activation of the G-protein subunit Galphas within forebrain neurons causes PKA-dependent alterations in fear conditioning and cortical Arc mRNA expression. Learn Mem 15: 75–83.
Kelly MP, Isiegas C, Cheung YF, Tokarczyk J, Yang X, Esposito MF et al (2007). Constitutive activation of Galphas within forebrain neurons causes deficits in sensorimotor gating because of PKA-dependent decreases in cAMP. Neuropsychopharmacology 32: 577–588.
Kelly MP, Logue SF, Brennan J, Day JP, Lakkaraju S, Jiang L et al (2010). Phosphodiesterase 11A in brain is enriched in ventral hippocampus and deletion causes psychiatric disease-related phenotypes. Proc Natl Acad Sci USA 107: 8457–8462.
Kelly MP, Stein JM, Vecsey CG, Favilla C, Yang X, Bizily SF et al (2009). Developmental etiology for neuroanatomical and cognitive deficits in mice overexpressing Galphas, a G-protein subunit genetically linked to schizophrenia. Mol Psychiatry 14: 398–415, 347.
Kelsoe J (2010) Method to Predict Response to Treatment for Psychiatric Illnesses vol. us20100233702 a1 (office, u. p. t., ed), p 1 The Regents of the University of California (Oakland, CA): USA.
Kogan JH, Frankland PW, Silva AJ (2000). Long-term memory underlying hippocampus-dependent social recognition in mice. Hippocampus 10: 47–56.
Laviola G, Terranova ML, Sedowofia K, Clayton R, Manning A (1994). A mouse model of early social interactions after prenatal drug exposure: a genetic investigation. Psychopharmacology (Berl) 113: 388–394.
Liebenberg N, Harvey BH, Brand L, Wegener G, Brink CB (2012). Chronic treatment with the phosphodiesterase type 5 inhibitors sildenafil and tadalafil display anxiolytic effects in Flinders Sensitive Line rats. Metab Brain Dis 27: 337–340.
Lukas M, Neumann ID (2013). Oxytocin and vasopressin in rodent behaviors related to social dysfunctions in autism spectrum disorders. Behav Brain Res 251: 85–94.
Luo HR, Wu GS, Dong C, Arcos-Burgos M, Ribeiro L, Licinio J et al (2009). Association of PDE11A global haplotype with major depression and antidepressant drug response. Neuropsychiatr Dis Treat 5: 163–170.
Mertens J, Wang QW, Kim Y, Yu DX, Pham S, Yang B et al (2015). Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature 527: 95–99.
Munger SD, Leinders-Zufall T, McDougall LM, Cockerham RE, Schmid A, Wandernoth P et al (2010). An olfactory subsystem that detects carbon disulfide and mediates food-related social learning. Curr Biol 20: 1438–1444.
Owen GR, Brenner EA (2012). Mapping molecular memory: navigating the cellular pathways of learning. Cell Mol Neurobiol 32: 919–941.
Pathak G, Ibrahim BA, McCarthy SA, Baker K, Kelly MP (2015). Amphetamine sensitization in mice is sufficient to produce both manic- and depressive-related behaviors as well as changes in the functional connectivity of corticolimbic structures. Neuropharmacology 95: 434–447.
Pathak G, Agostino MJ, Bishara K, Capell WR, Fisher JL, Hegde S et al (2016). PDE11A negatively regulates lithium responsivity. Mol Psych (under review).
Rosenberg T, Gal-Ben-Ari S, Dieterich DC, Kreutz MR, Ziv NE, Gundelfinger ED et al (2014). The roles of protein expression in synaptic plasticity and memory consolidation. Front Mol Neurosci 7: 86.
Schultz JE, Dunkern T, Gawlitta-Gorka E, Sorg G (2011). The GAF-tandem domain of phosphodiesterase 5 as a potential drug target. Handb Exp Pharmacol 204: 151–166.
Srivastava DP, Jones KA, Woolfrey KM, Burgdorf J, Russell TA, Kalmbach A et al (2012). Social, communication, and cortical structural impairments in Epac2-deficient mice. J Neurosci 32: 11864–11878.
Stoesz BM, Hare JF, Snow WM (2013). Neurophysiological mechanisms underlying affiliative social behavior: insights from comparative research. Neurosci Biobehav Rev 37: 123–132.
Tipton LA, Christensen L, Blacher J (2013). Friendship quality in adolescents with and without an intellectual disability. J Appl Res Intellect Disabil 26: 522–532.
Tseng KY, Chambers RA, Lipska BK (2009). The neonatal ventral hippocampal lesion as a heuristic neurodevelopmental model of schizophrenia. Behav Brain Res 204: 295–305.
Viero C, Shibuya I, Kitamura N, Verkhratsky A, Fujihara H, Katoh A et al (2010). REVIEW: Oxytocin: Crossing the bridge between basic science and pharmacotherapy. CNS Neurosci Ther 16: e138–e156.
Wong ML, Whelan F, Deloukas P, Whittaker P, Delgado M, Cantor RM et al (2006). Phosphodiesterase genes are associated with susceptibility to major depression and antidepressant treatment response. Proc Natl Acad Sci USA 103: 15124–15129.
Wood SK, Baez MA, Bhatnagar S, Valentino RJ (2009). Social stress-induced bladder dysfunction: potential role of corticotropin-releasing factor. Am J Physiol Regul Integr Comp Physiol 296: R1671–R1678.
Wrenn CC, Harris AP, Saavedra MC, Crawley JN (2003). Social transmission of food preference in mice: methodology and application to galanin-overexpressing transgenic mice. Behav Neurosci 117: 21–31.
Yang Y, Shu X, Liu D, Shang Y, Wu Y, Pei L et al (2012). EPAC null mutation impairs learning and social interactions via aberrant regulation of miR-124 and Zif268 translation. Neuron 73: 774–788.
Yuasa K, Kanoh Y, Okumura K, Omori K (2001a). Genomic organization of the human phosphodiesterase PDE11A gene. Evolutionary relatedness with other PDEs containing GAF domains. Eur J Biochem 268: 168–178.
Yuasa K, Kotera J, Fujishige K, Michibata H, Sasaki T, Omori K (2000). Isolation and characterization of two novel phosphodiesterase PDE11A variants showing unique structure and tissue-specific expression. J Biol Chem 275: 31469–31479.
Yuasa K, Ohgaru T, Asahina M, Omori K (2001b). Identification of rat cyclic nucleotide phosphodiesterase 11A (PDE11A): comparison of rat and human PDE11A splicing variants. Eur J Biochem 268: 4440–4448.
Acknowledgements
We would like to thank Joseph Meyers for technical support and Drs Susan and Christopher Wood and Dr Alexander McDonald for technical advice regarding immunofluorescence.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on the Neuropsychopharmacology website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Hegde, S., Capell, W., Ibrahim, B. et al. Phosphodiesterase 11A (PDE11A), Enriched in Ventral Hippocampus Neurons, is Required for Consolidation of Social but not Nonsocial Memories in Mice. Neuropsychopharmacol 41, 2920–2931 (2016). https://doi.org/10.1038/npp.2016.106
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2016.106
This article is cited by
-
PDE11A negatively regulates lithium responsivity
Molecular Psychiatry (2017)