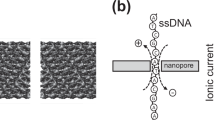
Abstract
Compared with conventional methods, single-molecule real-time (SMRT) DNA sequencing exhibits longer read lengths than conventional methods, less GC bias, and the ability to read DNA base modifications. However, reading DNA sequence from sub-nanogram quantities is impractical owing to inefficient delivery of DNA molecules into the confines of zero-mode waveguides—zeptolitre optical cavities in which DNA sequencing proceeds. Here, we show that the efficiency of voltage-induced DNA loading into waveguides equipped with nanopores at their floors is five orders of magnitude greater than existing methods. In addition, we find that DNA loading is nearly length-independent, unlike diffusive loading, which is biased towards shorter fragments. We demonstrate here loading and proof-of-principle four-colour sequence readout of a polymerase-bound 20,000-base-pair-long DNA template within seconds from a sub-nanogram input quantity, a step towards low-input DNA sequencing and mammalian epigenomic mapping of native DNA samples.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Flusberg, B. A. et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465 (2010).
Chaisson, M. J. P. et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature 517, 608–611 (2014).
Levene, M. J. et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299, 682–686 (2003).
Berlin, K. et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol. 33, 623–630 (2015).
Chaisson, M. J. P., Wilson, R. K. & Eichler, E. E. Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet. 16, 627–640 (2015).
Vilfan, I. D. et al. Analysis of RNA base modification and structural rearrangement by single-molecule real-time detection of reverse transcription. J. Nanobiotechnology 11, 8 (2013).
Jose, M. M.-M. et al. Cell investigation of nanostructures: zero-mode waveguides for plasma membrane studies with single molecule resolution. Nanotechnology 18, 195101 (2007).
Miyake, T. et al. Real-time imaging of single-molecule fluorescence with a zero-mode waveguide for the analysis of protein−protein interaction. Anal. Chem. 80, 6018–6022 (2008).
Uemura, S. et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature 464, 1012–1017 (2010).
Sandén, T. et al. A zeptoliter volume meter for analysis of single protein molecules. Nano Lett. 12, 370–375 (2012).
Richards, C. I. et al. Live-cell imaging of single receptor composition using zero-mode waveguide nanostructures. Nano Lett. 12, 3690–3694 (2012).
de Torres, J. et al. FRET enhancement in aluminum zero-mode waveguides. ChemPhysChem 16, 782–788 (2015).
Robertson, R. M., Laib, S. & Smith, D. E. Diffusion of isolated DNA molecules: dependence on length and topology. Proc. Natl Acad. Sci. USA 103, 7310–7314 (2006).
Pedone, D., Langecker, M., Abstreiter, G. & Rant, U. A pore−cavity−pore device to trap and investigate single nanoparticles and DNA molecules in a femtoliter compartment: confined diffusion and narrow escape. Nano Lett. 11, 1561–1567 (2011).
Liu, X., Skanata, M. M. & Stein, D. Entropic cages for trapping DNA near a nanopore. Nat. Commun. 6, 6222 (2015).
Han, J., Turner, S. W. & Craighead, H. G. Entropic trapping and escape of long DNA molecules at submicron size constriction. Phys. Rev. Lett. 83, 1688–1691 (1999).
Coupland, P. et al. Direct sequencing of small genomes on the Pacific Biosciences RS without library preparation. BioTechniques 53, 365–372 (2012).
Raley, C. et al. Preparation of next-generation DNA sequencing libraries from ultra-low amounts of input DNA: application to single-molecule, real-time (SMRT) sequencing on the Pacific Biosciences RS II. Preprint at bioRxivhttp://www.biorxiv.org/content/early/2014/03/25/003566 (2014).
Larkin, J. et al. Reversible positioning of single molecules inside zero-mode waveguides. Nano Lett. 14, 6023–6029 (2014).
Lundquist, P. M. et al. Parallel confocal detection of single molecules in real time. Opt. Lett. 33, 1026–1028 (2008).
Assad, O. N., Di Fiori, N., Squires, A. H. & Meller, A. Two color DNA barcode detection in photoluminescence suppressed silicon nitride nanopores. Nano Lett. 15, 745–752 (2015).
Sawafta, F. et al. Solid-state nanopores and nanopore arrays optimized for optical detection. Nanoscale 6, 6991–6996 (2014).
Sun, S., Rao, V. B. & Rossmann, M. G. Genome packaging in viruses. Curr. Opin. Struct. Biol. 20, 114–120 (2010).
Zhu, P. & Craighead, H. G. Zero-mode waveguides for single-molecule analysis. Annu. Rev. Biophys. 41, 269–293 (2012).
Wulfmeyer, T. et al. Structural organization of DNA in chlorella viruses. PLoS ONE 7, e30133 (2012).
Godfrey, J. E. & Eisenberg, H. The flexibility of low molecular weight double-stranded DNA as a function of length. II. Light scattering measurements and the estimation of persistence lengths from light scattering, sedimentation and viscosity. Biophys. Chem. 5, 301–318 (1976).
Wanunu, M. et al. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat. Nanotech. 5, 160–165 (2010).
Bell, N. A. W., Muthukumar, M. & Keyser, U. F. Translocation frequency of double-stranded DNA through a solid-state nanopore. Phys. Rev. E 93, 022401 (2016).
Freedman, K. J. et al. Nanopore sensing at ultra-low concentrations using single-molecule dielectrophoretic trapping. Nat. Commun. 7, 10217 (2016).
Grosberg, A. Y. & Rabin, Y. DNA capture into a nanopore: interplay of diffusion and electrohydrodynamics. J. Chem. Phys. 133, 165102 (2010).
Muthukumar, M. Theory of capture rate in polymer translocation. J. Chem. Phys. 132, 195101 (2010).
Stellwagen, N. C., Gelfi, C. & Righetti, P. G. The free solution mobility of DNA. Biopolymers 42, 687–703 (1997).
Wanunu, M. et al. DNA translocation governed by interactions with solid-state nanopores. Biophys. J. 95, 4716–4725 (2008).
Wanunu, M. et al. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat. Nanotech. 5, 807–814 (2010).
Zahid, O. K. et al. Sequence-specific recognition of microRNAs and other short nucleic acids with solid-state nanopores. Nano Lett. 16, 2033–2039 (2016).
Kowalczyk, S. W. & Dekker, C. Measurement of the docking time of a DNA molecule onto a solid-state nanopore. Nano Lett. 12, 4159–4163 (2012).
Yamazaki, H., Ito, S., Esashika, K. & Saiki, T. Optical observation of DNA motion during and immediately after nanopore translocation. Appl. Phys. Express 9, 017001 (2016).
Berndsen, Z. T. et al. Nonequilibrium dynamics and ultraslow relaxation of confined DNA during viral packaging. Proc. Natl Acad. Sci. USA 111, 8345–8350 (2014).
Wilchek, M. & Bayer, E. A. The avidin-biotin complex in bioanalytical applications. Anal. Biochem. 171, 1–32 (1988).
Srisa-Art, M., Dyson, E. C., deMello, A. J. & Edel, J. B. Monitoring of real-time streptavidin−biotin binding kinetics using droplet microfluidics. Anal. Chem. 80, 7063–7067 (2008).
Korlach, J. et al. Long, processive enzymatic DNA synthesis using 100% dye-labeled terminal phosphate-linked nucleotides. Nucleosides Nucleotides Nucleic Acids 27, 1072–1082 (2008).
Acknowledgements
We acknowledge Y.-C. Tsai, I. Vilfan, J. Hanes, R. Lam and M. McCauley for aid in sample preparation, as well as J. Sutin for assistance with the multimode fibre setup on our microscope. This work was supported by funding from the National Institutes of Health (HG006873 and HG009186, to M.W. and J.K.). This work was performed in part at the Cornell Nanoscale Facility, a member of the National Nanotechnology Infrastructure Network (NNIN), which is supported by the National Science Foundation (grant ECCS-1542081).
Author information
Authors and Affiliations
Contributions
J.L. and M.W. conceived and designed the experiments. J.L. and V.J. fabricated the NZMW devices. J.L., V.J. and R.Y.H. performed the experiments and analysed the data. R.Y.H. wrote the sequence analysis code. All authors wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.K. is a full-time employee at Pacific Biosciences, a company developing sequencing technologies.
Supplementary information
Supplementary information
Supplementary information (PDF 2598 kb)
Supplementary information
Supplementary Movie 1 (MP4 2504 kb)
Supplementary information
Supplementary Movie 2 (MP4 30563 kb)
Rights and permissions
About this article
Cite this article
Larkin, J., Henley, R., Jadhav, V. et al. Length-independent DNA packing into nanopore zero-mode waveguides for low-input DNA sequencing. Nature Nanotech 12, 1169–1175 (2017). https://doi.org/10.1038/nnano.2017.176
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2017.176
This article is cited by
-
Nanopore DNA sequencing technologies and their applications towards single-molecule proteomics
Nature Chemistry (2024)
-
MoS2 nanopore identifies single amino acids with sub-1 Dalton resolution
Nature Communications (2023)
-
Cancer nanotechnology: current status and perspectives
Nano Convergence (2021)
-
Solid-State Nanopore for Molecular Detection
International Journal of Precision Engineering and Manufacturing (2021)
-
Genetic Analyzer Nanofor 05 as a Measuring Instrument for DNA Sequencing
Measurement Techniques (2021)