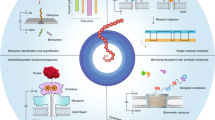
Abstract
Nanopore DNA sequencing offers a new paradigm owing to its extensive potential for long-read, high-throughput detection of nucleotide modification and direct RNA sequencing. Given the remarkable advances in protein nanopore sequencing technology, there is still a strong enthusiasm in exploring alternative nanopore-sequencing techniques, particularly those based on a solid-state nanopore using a semiconductor material. Since solid-state nanopores provide superior material robustness and large-scale integrability with on-chip electronics, they have the potential to surpass the limitations of their biological counterparts. However, there are key technical challenges to be addressed: the creation of an ultrasmall nanopore, fabrication of an ultrathin membrane, control of the ultrafast DNA speed and detection of four nucleotides. Extensive research efforts have been devoted to resolving these issues over the past two decades. In this review, we briefly introduce recent updates regarding solid-state nanopore technologies towards DNA sequencing. It can be envisioned that emerging technologies will offer a brand new future in DNA-sequencing technology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA, et al. DNA sequencing at 40: past, present and future. Nature. 2017;550:345–53.
Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–51.
Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–53.
Deamer D, Akeson M, Branton D. Three decades of nanopore sequencing. Nat Biotechnol. 2016;34:518–24.
Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA 1996;93:13770–3.
Wang S, Zhao Z, Haque F, Guo P. Engineering of protein nanopores for sequencing, chemical or protein sensing and disease diagnosis. Curr Opin Biotechnol. 2018;51:80–9.
Editorial. The long view on sequencing. Nat Biotechnol. 2018;36:287.
Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018;36:338–45.
Depledge DP, Srinivas KP, Sadaoka T, Bready D, Mori Y, Placantonakis DG, et al. Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen. Nat Commun. 2019;10:754.
Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14:407–10.
Dekker C. Solid-state nanopores. Nat Nanotechnol. 2007;2:209–15.
Lindsay S. The promises and challenges of solid-state sequencing. Nat Nanotechnol. 2016;11:109–11.
Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–9.
Iqbal SM, Bashir R. Nanopores: sensing and fundamental biological interactions. Heidelberg: Springer; 2011.
Edel JB, Albrecht T. Engineered nanopores for bioanalytical applications. Amsterdam, NL: Elsevier Science; 2013.
Wanunu M. Nanopores: a journey towards DNA sequencing. Phys Life Rev. 2012;9:125–58.
Carson S, Wanunu M. Challenges in DNA motion control and sequence readout using nanopore devices. Nanotechnology. 2015;26:074004.
Wang Y, Yang Q, Wang Z. The evolution of nanopore sequencing. Front Genet 2014;5:449.
Agah S, Zheng M, Pasquali M, Kolomeisky AB. DNA sequencing by nanopores: advances and challenges. J Phys D. 2016;49:413001.
Tang Z, Zhang D, Cui W, Zhang H, Pang W, Duan X. Fabrications, applications and challenges of solid-state nanopores: a mini review. Nanomater Nanotechnol 2016;6:35.
Lee K, Park KB, Kim HJ, Yu JS, Chae H, Kim HM, et al. Recent progress in solid-state nanopores. Adv Mater. 2018;30:e1704680.
Yuan Z, Wang C, Yi X, Ni Z, Chen Y, Li T. Solid-state nanopore. Nanoscale Res Lett. 2018;13:56.
Pungetmongkol P. Speculation of nano-gap sensor for DNA sequencing technology: a review on synthetic nanopores. Eng J. 2018;22:229–50.
Huang S, Romero-Ruiz M, Castell OK, Bayley H, Wallace MI. High-throughput optical sensing of nucleic acids in a nanopore array. Nat Nanotechnol. 2015;10:986.
Howorka S, Cheley S, Bayley H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nat Biotechnol. 2001;19:636–9.
Pennisi E. Search for pore-fection. Science. 2012;336:534.
Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30:349–53.
Laszlo AH, Derrington IM, Ross BC, Brinkerhoff H, Adey A, Nova IC, et al. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32:829–33.
Meller A. Dynamics of polynucleotide transport through nanometre-scale pores. J Phys Condens Matter 2003;15:R581.
Akahori R, Haga T, Hatano T, Yanagi I, Ohura T, Hamamura H, et al. Slowing single-stranded DNA translocation through a solid-state nanopore by decreasing the nanopore diameter. Nanotechnology. 2014;25:275501.
Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Fabrication of solid-state nanopores with single-nanometre precision. Nat Mater. 2003;2:537–40.
Larkin J, Henley R, Bell DC, Cohen-Karni T, Rosenstein JK, Wanunu M. Slow DNA transport through nanopores in hafnium oxide membranes. ACS Nano 2013;7:10121–8.
Kwok H, Briggs K, Tabard-Cossa V. Nanopore fabrication by controlled dielectric breakdown. PLoS ONE. 2014;9:e92880.
Yanagi I, Akahori R, Hatano T, Takeda K. Fabricating nanopores with diameters of sub-1 nm to 3 nm using multilevel pulse-voltage injection. Sci Rep. 2014;4:5000.
Briggs K, Kwok H, Tabard-Cossa V. Automated fabrication of 2-nm solid-state nanopores for nucleic acid analysis. Small 2014;10:2077–86.
Goto Y, Yanagi I, Matsui K, Yokoi T, Takeda K. Integrated solid-state nanopore platform for nanopore fabrication via dielectric breakdown, DNA-speed deceleration and noise reduction. Sci Rep. 2016;6:31324.
Tahvildari R, Beamish E, Tabard-Cossa V, Godin M. Integrating nanopore sensors within microfluidic channel arrays using controlled breakdown. Lab Chip. 2015;15:1407–11.
Yanagi I, Akahori R, Aoki M, Harada K, Takeda K. Multichannel detection of ionic currents through two nanopores fabricated on integrated Si3N4 membranes. Lab Chip. 2016;16:3340–50.
Pud S, Verschueren DV, Vukovic N, Plesa C, Jonsson M, Dekker C. Self-aligned plasmonic nanopores by optically controlled dielectric breakdown. Nano Lett. 2015;15:7112.
Gilboa T, Zrehen A, Girsault A, Meller A. Optically-monitored nanopore fabrication using a focused laser beam. Sci Rep. 2018;8:9765.
Briggs K, Charron M, Kwok H, Le T, Chahal S, Bustamante J, et al. Kinetics of nanopore fabrication during controlled breakdown of dielectric membranes in solution. Nanotechnology. 2015;26:084004.
Matsui K, Yanagi I, Goto Y, Takeda K. Prevention of dielectric breakdown of nanopore membranes by charge neutralization. Sci Rep. 2015;5:17819.
Ying C, Zhang Y, Feng Y, Zhou D, Wang D, Xiang Y, et al. 3D nanopore shape control by current-stimulus dielectric breakdown. Appl Phys Lett. 2016;109:063105.
Arcadia CE, Reyes CC, Rosenstein JK. In-situ nanopore fabrication and single-molecule sensing with microscale liquid contacts. ACS Nano 2017;11:4907.
Yanagi I, Fujisaki K, Hamamura H, Takeda K. Thickness-dependent dielectric breakdown and nanopore creation on sub-10-nm-thick SiN membranes in solution. J Appl Phys. 2017;121:045301.
Carlsen AT, Briggs K, Hall AR, Tabard-Cossa V. Solid-state nanopore localization by controlled breakdown of selectively thinned membranes. Nanotechnology. 2017;28:085304.
Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LM, et al. DNA translocation through graphene nanopores. Nano Lett. 2010;10:3163–7.
Liu K, Feng J, Kis A, Radenovic A. Atomically thin molybdenum disulfide nanopores with high sensitivity for DNA translocation. ACS Nano 2014;8:2504–11.
Liu S, Lu B, Zhao Q, Li J, Gao T, Chen Y, et al. Boron nitride nanopores: highly sensitive DNA single-molecule detectors. Adv Mater. 2013;25:4549–54.
Heerema SJ, Dekker C. Graphene nanodevices for DNA sequencing. Nat Nanotechnol. 2016;11:127.
Venta K, Shemer G, Puster M, Rodriguez-Manzo JA, Balan A, Rosenstein JK, et al. Differentiation of short, single-stranded DNA homopolymers in solid-state nanopores. ACS Nano 2013;7:4629.
Carlsen AT, Zahid OK, Ruzicka J, Taylor EW, Hall AR. Interpreting the conductance blockades of DNA translocations through solid-state nanopores. ACS Nano 2014;8:4754–60.
Lee M-H, Kumar A, Park K-B, Cho S-Y, Kim H-M, Lim M-C, et al. A low-noise solid-state nanopore platform based on a highly insulating substrate. Sci Rep. 2014;4:7448.
Yanagi I, Ishida T, Fujisaki K, Takeda K. Fabrication of 3-nm-thick Si3N4 membranes for solid-state nanopores using the poly-Si sacrificial layer process. Sci Rep. 2015;5:14656.
Wanunu M, Dadosh T, Ray V, Jin J, McReynolds L, Drndić M. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat Nanotechnol. 2010;5:807.
Yamazaki H, Hu R, Zhao Q, Wanunu M. Photothermally assisted thinning of silicon nitride membranes for ultrathin asymmetric nanopores. ACS Nano 2018;12:12472–81.
Wanunu M, Sutin J, McNally B, Chow A, Meller A. DNA translocation governed by interactions with solid-state nanopores. Biophys J. 2008;95:4716–25.
Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nat Nanotechnol. 2011;6:615–24.
Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;5:1734–7.
Kowalczyk SW, Wells DB, Aksimentiev A, Dekker C. Slowing down DNA translocation through a nanopore in lithium chloride. Nano Lett. 2012;12:1038–44.
Verschueren DV, Jonsson MP, Dekker C. Temperature dependence of DNA translocations through solid-state nanopores. Nanotechnology. 2015;26:234004.
Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat Nanotechnol. 2009;5:160.
Squires AH, Hersey JS, Grinstaff MW, Meller A. A nanopore-nanofiber mesh biosensor to control DNA translocation. J Am Chem Soc 2013;135:16304–7.
Goto Y, Haga T, Yanagi I, Yokoi T, Takeda K. Deceleration of single-stranded DNA passing through a nanopore using a nanometre-sized bead structure. Sci Rep. 2015;5:16640.
Tang Z, Liang Z, Lu B, Li J, Hu R, Zhao Q, et al. Gel mesh as “brake” to slow down DNA translocation through solid-state nanopores. Nanoscale 2015;7:13207–14.
Yoshida H, Goto Y, Akahori R, Tada Y, Terada S, Komura M, et al. Slowing the translocation of single-stranded DNA by using nano-cylindrical passage self-assembled by amphiphilic block copolymers. Nanoscale 2016;8:18270–6.
Keyser UF, Koeleman BN, van Dorp S, Krapf D, Smeets RMM, Lemay SG, et al. Direct force measurements on DNA in a solid-state nanopore. Nat Phys. 2006;2:473–7.
Nelson EM, Li H, Timp G. Direct, concurrent measurements of the forces and currents affecting DNA in a nanopore with comparable topography. ACS Nano 2014;8:5484–93.
Akahori R, Yanagi I, Goto Y, Harada K, Yokoi T, Takeda K. Discrimination of three types of homopolymers in single-stranded DNA with solid-state nanopores through external control of the DNA motion. Sci Rep. 2017;7:9073.
Lu B, Albertorio F, Hoogerheide DP, Golovchenko JA. Origins and consequences of velocity fluctuations during DNA passage through a nanopore. Biophys J. 2011;101:70–9.
Feng J, Liu K, Bulushev RD, Khlybov S, Dumcenco D, Kis A, et al. Identification of single nucleotides in MoS2 nanopores. Nat Nanotechnol. 2015;10:1070.
Goto Y, Yanagi I, Matsui K, Yokoi T, Takeda K. Identification of four single-stranded DNA homopolymers with a solid-state nanopore in alkaline CsCl solution. Nanoscale 2018;10:20844–50.
Derrington IM, Butler TZ, Collins MD, Manrao E, Pavlenok M, Niederweis M, et al. Nanopore DNA sequencing with MspA. Proc Natl Acad Sci USA 2010;107:16060–5.
Comer J, Aksimentiev A. DNA sequence-dependent ionic currents in ultra-small solid-state nanopores. Nanoscale 2016;8:9600–13.
Clarke J, Wu H-C, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol. 2009;4:265–70.
Ayub M, Hardwick SW, Luisi BF, Bayley H. Nanopore-based identification of individual nucleotides for direct RNA sequencing. Nano Lett. 2013;13:6144–50.
Kumar S, Tao C, Chien M, Hellner B, Balijepalli A, Robertson JWF, et al. PEG-labeled nucleotides and nanopore detection for single molecule DNA sequencing by synthesis. Sci Rep. 2012;2:684.
Stranges PB, Palla M, Kalachikov S, Nivala J, Dorwart M, Trans A, et al. Design and characterization of a nanopore-coupled polymerase for single-molecule DNA sequencing by synthesis on an electrode array. Proc Natl Acad Sci USA 2016;113:E6749–56.
Lee JW. Nanoelectrode-gated detection of individual molecules with potential for rapid DNA sequencing. Solid State Phenom 2007;121-123:1379–86.
Tsutsui M, Taniguchi M, Kawai T. Fabrication of 0.5 nm electrode gaps using self-breaking technique. Appl Phys Lett. 2008;93:163115.
Tsutsui M, Taniguchi M, Yokota K, Kawai T. Identifying single nucleotides by tunnelling current. Nat Nanotechnol. 2010;5:286–90.
Ohshiro T, Matsubara K, Tsutsui M, Furuhashi M, Taniguchi M, Kawai T. Single-molecule electrical random resequencing of DNA and RNA. Sci Rep. 2012;2:501.
Ohshiro T, Tsutsui M, Yokota K, Taniguchi M. Quantitative analysis of DNA with single-molecule sequencing. Sci Rep. 2018;8:8517.
Taniguchi M. Selective multidetection using nanopores. Anal Chem. 2015;87:188–99.
Acknowledgements
The authors would like to express the utmost thanks to all coworkers for their dedication to Hitachi’s solid-state nanopore DNA sequencer project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Financial support for this work was provided by Hitachi, Ltd. Y.G., R.A., I.Y., and K.T. are current employees of Hitachi, Ltd.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Goto, Y., Akahori, R., Yanagi, I. et al. Solid-state nanopores towards single-molecule DNA sequencing. J Hum Genet 65, 69–77 (2020). https://doi.org/10.1038/s10038-019-0655-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s10038-019-0655-8