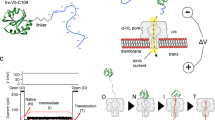
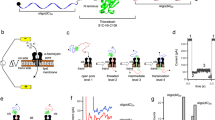
Abstract
Cells are divided into compartments and separated from the environment by lipid bilayer membranes. Essential molecules are transported back and forth across the membranes. We have investigated how folded proteins use narrow transmembrane pores to move between compartments. During this process, the proteins must unfold. To examine co-translocational unfolding of individual molecules, we tagged protein substrates with oligonucleotides to enable potential-driven unidirectional movement through a model protein nanopore, a process that differs fundamentally from extension during force spectroscopy measurements. Our findings support a four-step translocation mechanism for model thioredoxin substrates. First, the DNA tag is captured by the pore. Second, the oligonucleotide is pulled through the pore, causing local unfolding of the C terminus of the thioredoxin adjacent to the pore entrance. Third, the remainder of the protein unfolds spontaneously. Finally, the unfolded polypeptide diffuses through the pore into the recipient compartment. The unfolding pathway elucidated here differs from those revealed by denaturation experiments in solution, for which two-state mechanisms have been proposed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Van der Laan, M. et al. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nature Cell Biol. 9, 1152–1159 (2007).
Krayl, M., Lim, J. H., Martin, F., Guiard, B. & Voos, W. A cooperative action of the ATP-dependent import motor complex and the inner membrane potential drives mitochondrial preprotein import. Mol. Cell Biol. 27, 411–425 (2007).
Huang, S., Ratliff, K. S. & Matouschek, A. Protein unfolding by the mitochondrial membrane potential. Nature Struct. Biol. 9, 301–307 (2002).
Collier, R. J. Membrane translocation by anthrax toxin. Mol. Aspects Med. 30, 413–422 (2009).
Papadakos, G., Wojdyla, J. A. & Kleanthous, C. Nuclease colicins and their immunity proteins. Q. Rev. Biophys. 45, 57–103 (2012).
Martin, A., Baker, T. A. & Sauer, R. T. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nature Struct. Mol. Biol. 15, 1147–1151 (2008).
Maillard, R. A. et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell 145, 459–469 (2011).
Tian, P. & Andricioaei, I. Repetitive pulling catalyzes co-translocational unfolding of barnase during import through a mitochondrial pore. J. Mol. Biol. 350, 1017–1034 (2005).
Makarov, D. E. Computer simulations and theory of protein translocation. Acc. Chem. Res. 42, 281–289 (2009).
Muthukumar, M. Polymer Translocation (CRC, 2011).
Rincon-Restrepo, M., Mikhailova, E., Bayley, H. & Maglia, G. Controlled translocation of individual DNA molecules through protein nanopores with engineered molecular brakes. Nano Lett. 11, 746–750 (2011).
Krantz, B. A. et al. A phenylalanine clamp catalyzes protein translocation through the anthrax toxin pore. Science 309, 777–781 (2005).
Basilio, D., Juris, S. J., Collier, R. J. & Finkelstein, A. Evidence for a proton–protein symport mechanism in the anthrax toxin channel. J. Gen. Physiol. 133, 307–314 (2009).
Thoren, K. L., Worden, E. J., Yassif, J. M. & Krantz, B. A. Lethal factor unfolding is the most force-dependent step of anthrax toxin translocation. Proc. Natl Acad. Sci. USA 106, 21555–21560 (2009).
Basilio, D., Jennings-Antipov, L. D., Jakes, K. S. & Finkelstein, A. Trapping a translocating protein within the anthrax toxin channel: implications for the secondary structure of permeating proteins. J. Gen. Physiol. 137, 343–356 (2011).
Basilio, D., Kienker, P. K., Briggs, S. W. & Finkelstein, A. A kinetic analysis of protein transport through the anthrax toxin channel. J. Gen. Physiol. 137, 521–531 (2011).
Simon, S. M., Peskin, C. S. & Oster, G. F. What drives the translocation of proteins? Proc. Natl Acad. Sci. USA 89, 3770–3774 (1992).
Krantz, B. A., Finkelstein, A. & Collier, R. J. Protein translocation through the anthrax toxin transmembrane pore is driven by a proton gradient. J. Mol. Biol. 355, 968–979 (2006).
Pentelute, B. L., Sharma, O. & Collier, R. J. Chemical dissection of protein translocation through the anthrax toxin pore. Angew Chem. Int. Ed. 50, 2294–2296 (2011).
Wigelsworth, D. J. et al. Binding stoichiometry and kinetics of the interaction of a human anthrax toxin receptor, CMG2, with protective antigen. J. Biol. Chem. 279, 23349–23356 (2004).
Chacinska, A., Koehler, C. M., Milenkovic, D., Lithgow, T. & Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009).
Oukhaled, G. et al. Unfolding of proteins and long transient conformations detected by single nanopore recording. Phys. Rev. Lett. 98, 158101 (2007).
Talaga, D. S. & Li, J. Single-molecule protein unfolding in solid state nanopores. J. Am. Chem. Soc. 131, 9287–9297 (2009).
Payet, L. et al. Thermal unfolding of proteins probed at the single molecule level using nanopores. Anal. Chem. 84, 4071–4076 (2012).
Pastoriza-Gallego, M. et al. Dynamics of unfolded protein transport through an aerolysin pore. J. Am. Chem. Soc. 133, 2923–2931 (2011).
Merstorf, C. et al. Wild type, mutant protein unfolding and phase transition detected by single-nanopore recording. ACS Chem. Biol. 7, 652–658 (2012).
Stefureac, R. I., Waldner, L., Howard, P. & Lee, J. S. Nanopore analysis of a small 86-residue protein. Small 4, 59–63 (2008).
Pey, A. L. et al. Engineering proteins with tunable thermodynamic and kinetics stabilities. Proteins 71, 165–174 (2008).
Rodriguez-Larrea, D. et al. Role of conservative mutations in protein multi-property adaption. Biochem. J. 429, 243–249 (2010).
Henrickson, S. E., Misakian, M., Robertson, B. & Kasianowicz, J. J. Driven DNA transport into an asymmetric nanometer-scale pore. Phys. Rev. Lett. 85, 3057–3060 (2000).
Rabin, Y. & Tanaka, M. DNA in nanopores: counterion condensation and coion depletion. Phys. Rev. Lett. 94, 148103 (2005).
Zhang, J. & Shklovskii, B. I. Effective charge and free energy of DNA inside an ion channel. Phys. Rev. E 75, 021906 (2007).
Dyson, H. J., Jeng, M-F., Model, P. & Holmgren, A. Characterization by 1H NMR of a C32S, C35S double mutant of Escherichia coli thioredoxin demonstrates its resemblance to the reduced wild type protein. FEBS Lett. 339, 11–17 (1994).
Holmgren, A. Thioredoxin structure and mechanism: conformational changes on oxidation of the active site sulfhydryls to a disulfide. Structure 3, 239–343 (1995).
Howorka, S., Cheley, S. & Bayley, H. Sequence-specific detection of individual DNA strands using engineered nanopores. Nature Biotechnol. 19, 636–639 (2001).
Maglia, G., Heron, A. J., Stoddart, D., Japrung, D. & Bayley, H. Analysis of single nucleic acid molecules with protein nanopores. Methods Enzymol. 475, 591–623 (2010).
Maglia, G., Restrepo, M. R., Mikhailova, E. & Bayley, H. Enhanced translocation of single DNA molecules through α-hemolysin nanopores by manipulation of internal charge. Proc. Natl Acad. Sci. USA 105, 19720–19725 (2008).
Stoddart, D., Heron, A. J., Mikhailova, E., Maglia, G. & Bayley, H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc. Natl Acad. Sci. USA 106, 7702–7707 (2009).
Japrung, D., Henricus, M., Li, Q., Maglia, G. & Bayley, H. Urea facilitates the translocation of single-stranded DNA and RNA through the α-hemolysin nanopore. Biophys. J. 98, 1856–1863 (2010).
Bennion, B. J. & Daggett, V. The molecular basis for the chemical denaturation of proteins by urea. Proc. Natl Acad. Sci. USA 100, 5142–5147 (2003).
Myers, J. K., Pace, C. N. & Scholtz, J. M. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 4, 2138–2148 (1995).
Rodriguez-Larrea, D., Ibarra-Molero, B. & Sanchez-Ruiz, J. M. Energetic and structural consequences of desolvation/solvation barriers to protein folding/unfolding assessed from experimental unfolding rates. Biophys. J. 91, L48–L50 (2006).
Muñoz, V. & Sanchez-Ruiz, J. M. Exploring protein-folding ensembles: a variable-barrier model for the analysis of equilibrium unfolding experiments. Proc. Natl Acad. Sci. USA 101, 17646–17651 (2004).
Bayley, H. et al. in Single Molecules and Nanotechnology (eds Rigler, R. & Vogel, H.) Ch. 10, 251–277 (Springer, 2008).
Schlierf, M., Li, H. & Fernandez, J. M. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc. Natl Acad. Sci. USA 101, 7299–7304 (2004).
Mathé, J., Visram, H., Viasnoff, V., Rabin, Y. & Meller, A. Nanopore unzipping of individual DNA hairpin molecules. Biophys. J. 87, 3205–3212 (2004).
Wiggin, M., Tropini, C., Tabard-Cossa, V., Jetha, N. N. & Marziali, A. Nonexponential kinetics of DNA escape from α-hemolysin nanopores. Biophys. J. 95, 5317–5323 (2008).
Matouschek, A., Pfanner, N. & Voos, W. Protein unfolding by mitochondria. The Hsp70 motor. EMBO Rep. 1, 404–410 (2000).
Acknowledgements
The authors thank E. Mikhailova for the αHL protein prepared by in vitro transcription and translation. D.R-L. is a recipient of an EMBO Long-Term Fellowship. This work was also supported by a grant from Oxford Nanopore Technologies.
Author information
Authors and Affiliations
Contributions
D.R-L. and H.B. planned the research. D.R-L. perfomed the experiments and data analysis. D.R-L. and H.B. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 1132 kb)
Rights and permissions
About this article
Cite this article
Rodriguez-Larrea, D., Bayley, H. Multistep protein unfolding during nanopore translocation. Nature Nanotech 8, 288–295 (2013). https://doi.org/10.1038/nnano.2013.22
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2013.22
This article is cited by
-
Peptide sequencing based on host–guest interaction-assisted nanopore sensing
Nature Methods (2024)
-
Enzyme-less nanopore detection of post-translational modifications within long polypeptides
Nature Nanotechnology (2023)
-
Study on the controllability of the fabrication of single-crystal silicon nanopores/nanoslits with a fast-stop ionic current-monitored TSWE method
Microsystems & Nanoengineering (2023)
-
Unidirectional single-file transport of full-length proteins through a nanopore
Nature Biotechnology (2023)
-
Polymer Translocation
Chinese Journal of Polymer Science (2023)