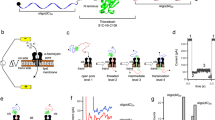
Abstract
Nanopores have recently been used to identify and fingerprint proteins. However, because proteins, unlike DNA, do not have a uniform charge, the electrophoretic force cannot in general be used to translocate or linearize them. Here we show that the introduction of sets of charges in the lumen of the CytK nanopore spaced by ~1 nm creates an electroosmotic flow that induces the unidirectional transport of unstructured natural polypeptides against a strong electrophoretic force. Molecular dynamics simulations indicate that this electroosmotic-dominated force has a strength of ~20 pN at −100 mV, which is similar to the electric force on single-stranded DNA. Unfolded polypeptides produce current signatures as they traverse the nanopore, which may be used to identify proteins. This approach can be used to translocate and stretch proteins for enzymatic and non-enzymatic protein identification and sequencing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data and the setup files for the simulations can be found at https://doi.org/10.5281/zenodo.8123395. Source data are provided with this paper.
References
Gu, L.-Q., Cheley, S. & Bayley, H. Electroosmotic enhancement of the binding of a neutral molecule to a transmembrane pore. Proc. Natl. Acad. Sci. USA 100, 15498–15503 (2003).
Huang, G., Willems, K., Soskine, M., Wloka, C. & Maglia, G. Electro-osmotic capture and ionic discrimination of peptide and protein biomarkers with FraC nanopores. Nat. Commun. 8, 935 (2017).
Huang, G. et al. Electro-osmotic vortices promote the capture of folded proteins by PlyAB nanopores. Nano Lett. 20, 3819–3827 (2020).
Asandei, A. et al. Electroosmotic trap against the electrophoretic force near a protein nanopore reveals peptide dynamics during capture and translocation. ACS Appl. Mater. Interfaces 8, 13166–13179 (2016).
Willems, K. et al. Engineering and modeling the electrophoretic trapping of a single protein inside a nanopore. ACS Nano 13, 9980–9992 (2019).
Gubbiotti, A. et al. Electroosmosis in nanopores: computational methods and technological applications. Adv. Phys. X 7, 2036638 (2022).
Brinkerhoff, H., Kang, A. S. W., Liu, J., Aksimentiev, A. & Dekker, C. Multiple rereads of single proteins at single–amino acid resolution using nanopores. Science 374, 1509–1513 (2021).
Yan, S. et al. Single molecule ratcheting motion of peptides in a Mycobacterium smegmatis porin A (MspA) nanopore. Nano Lett. 21, 6703–6710 (2021).
Chen, Z. et al. Controlled movement of ssDNA conjugated peptide through Mycobacterium smegmatis porin A (MspA) pore by a helicase motor for peptide sequencing application. Chem. Sci. 12, 15750–15756 (2021).
Rosen, C. B., Rodriguez-Larrea, D. & Bayley, H. Single-molecule site-specific detection of protein phosphorylation with a nanopore. Nat. Biotechnol. 32, 179–181 (2014).
Yu, L. et al. Unidirectional single-file transport of full-length proteins through a nanopore. Nat. Biotechnol. 41,1130–1139 (2023).
Biesemans, A., Soskine, M. & Maglia, G. A protein rotaxane controls the translocation of proteins across a ClyA nanopore. Nano Lett. 15, 6076–6081 (2015).
Aksimentiev, A. & Schulten, K. Imaging α-hemolysin with molecular dynamics: ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys. J. 88, 3745–3761 (2005).
Bonome, E. L., Cecconi, F. & Chinappi, M. Electroosmotic flow through an α-hemolysin nanopore. Microfluid. Nanofluid. 21,96 (2017).
Bétermier, F. et al. Single-sulfur atom discrimination of polysulfides with a protein nanopore for improved batteries. Commun. Mater. 1, 59 (2020).
Di Muccio, G., Morozzo della Rocca, B. & Chinappi, M. Geometrically induced selectivity and unidirectional electroosmosis in uncharged nanopores. ACS Nano 16, 8716–8728 (2022).
Raffy, S., Sassoon, N., Hofnung, M. & Betton, J.-M. Tertiary structure-dependence of misfolding substitutions in loops of the maltose-binding protein. Protein Sci. 7, 2136–2142 (1998).
Fonin, A. V. et al. Spectral characteristics of the mutant form GGBP/H152C of D-glucose/D-galactose-binding protein labeled with fluorescent dye BADAN: influence of external factors. PeerJ 2, e275 (2014).
Ohmae, E., Sasaki, Y. & Gekko, K. Effects of five-tryptophan mutations on structure, stability and function of Escherichia coli dihydrofolate reductase. J. Biochem. 130, 439–447 (2001).
Japrung, D., Henricus, M., Li, Q. H., Maglia, G. & Bayley, H. Urea facilitates the translocation of single-stranded DNA and RNA through the α-hemolysin nanopore. Biophys. J. 98, 1856–1863 (2010).
Pastoriza-Gallego, M. et al. Evidence of unfolded protein translocation through a protein nanopore. ACS Nano 8, 11350–11360 (2014).
Oukhaled, G. et al. Unfolding of proteins and long transient conformations detected by single nanopore recording. Phys. Rev. Lett. 98, 158101 (2007).
Pastoriza-Gallego, M. et al. Dynamics of unfolded protein transport through an aerolysin pore. J. Am. Chem. Soc. 133, 2923–2931 (2011).
Rodriguez-Larrea, D. & Bayley, H. Multistep protein unfolding during nanopore translocation. Nat. Nanotechnol. 8, 288–295 (2013).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Bitinaite, J. et al. USER friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res. 35, 1992–2002 (2007).
Cavaleiro, A. M., Kim, S. H., Seppälä, S., Nielsen, M. T. & Nørholm, M. H. H. Accurate DNA assembly and genome engineering with optimized uracil excision cloning. ACS Synth. Biol. 4, 1042–1046 (2015).
Nørholm, M. H. A mutant Pfu DNA polymerase designed for advanced uracil-excision DNA engineering. BMC Biotechnol. 10, 21 (2010).
Zhang, S. et al. Bottom-up fabrication of a proteasome–nanopore that unravels and processes single proteins. Nat. Chem. 13, 1192–1199 (2021).
Maglia, G., Heron, A. J. J. A. J., Stoddart, D., Japrung, D. & Bayley, H. Analysis of single nucleic acid molecules with protein nanopores. Methods Enzymol. 475, 591–623 (2010).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Tanaka, Y. et al. 2-Methyl-2,4-pentanediol induces spontaneous assembly of staphylococcal α-hemolysin into heptameric pore structure. Protein Sci. 20, 448–456 (2011).
Lambey, P. et al. Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins. eLife 11, e72555 (2022).
Nocadello, S. et al. Crystal structures of the components of the Staphylococcus aureus leukotoxin ED. Acta Crystallogr. Sect. D Struct. Biol. 72, 113–120 (2016).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Pettersen, E. F. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–12 (2004).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Yoo, J. & Aksimentiev, A. Improved parametrization of Li+, Na+, K+, and Mg2+ Ions for all-atom molecular dynamics simulations of nucleic acid systems. J. Phys. Chem. Lett. 3, 45–50 (2012).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Miyamoto, S. & Kollman, P. A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992).
Andersen, H. C. Rattle: a ‘velocity’ version of the shake algorithm for molecular dynamics calculations. J. Comput. Phys. 52, 24–34 (1983).
Martyna, G. J., Tobias, D. J. & Klein, M. L. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 (1994).
Gumbart, J., Khalili-Araghi, F., Sotomayor, M. & Roux, B. Constant electric field simulations of the membrane potential illustrated with simple systems. Biochim. Biophys. Acta 1818, 294–302 (2012).
Crozier, P. S., Henderson, D., Rowley, R. L. & Busath, D. D. Model channel ion currents in NaCl-extended simple point charge water solution with applied-field molecular dynamics. Biophys. J. 81, 3077–3089 (2001).
Acknowledgements
A.S. and G.M. acknowledge NWO-VICI grant 192.068 and NIH grant 1086554. M.C. and B.M.d.R acknowledge supercomputer time provided through the s1178 Production Grant from the Swiss National Supercomputing Centre (CSCS). M.C and B.M.d.R thank G. Di Muccio for useful discussions.
Author information
Authors and Affiliations
Contributions
G.M. and A.S. designed the research. A.S. carried out the experimental research. M.T. made the electrostatic maps of the nanopores. B.M.d.R. and M.C. carried out the simulations. All authors contributed to the writing of the manuscript and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. G.M. is a founder, director and shareholder of Portal Biotech Limited, a company engaged in the development of nanopore technologies. This work was not supported by Portal Biotech Limited.
Peer review
Peer review information
Nature Biotechnology thanks Henry Brinkerhoff and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–28 and Supplementary Tables 1–7
Supplementary Tables
This file contains the following tables: Supplementary Table 1: oligonucleotides (primers) used to generate the mutant CytK nanopores; Supplementary Table 2: oligonucleotides (primers) to generate the aerolysin mutant; Supplementary Table 3: oligonucleotides (primers) to generate the substrates; and Supplementary Table 4: the chemicals and reagents used in this study and their corresponding suppliers and catalog/article numbers.
Supplementary Data
Source data for Supplementary Fig. 28. Unprocessed SDS-PAGE of the substrates (proteins that were translocated across the nanopores).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sauciuc, A., Morozzo della Rocca, B., Tadema, M.J. et al. Translocation of linearized full-length proteins through an engineered nanopore under opposing electrophoretic force. Nat Biotechnol (2023). https://doi.org/10.1038/s41587-023-01954-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41587-023-01954-x
This article is cited by
-
Nanopore DNA sequencing technologies and their applications towards single-molecule proteomics
Nature Chemistry (2024)
-
Unclogged pores: designer channels for protein translocation
Communications Biology (2024)
-
Engineered nanopores for exopeptidase protein sequencing
Nature Methods (2024)
-
Real-time detection of 20 amino acids and discrimination of pathologically relevant peptides with functionalized nanopore
Nature Methods (2024)