Abstract
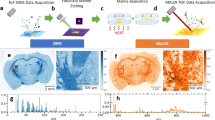
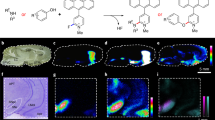
As large genomic and proteomic datasets are generated from homogenates of various tissues, the need for information on the spatial localization of their encoded products has become more pressing. Matrix-assisted laser desorption-ionization (MALDI) imaging mass spectrometry (IMS) offers investigators the means with which to unambiguously study peptides and proteins with molecular specificity, and to determine their distribution in two and three dimensions1,2. In the past few years, several parameters have been optimized for IMS, including sample preparation, matrix application and instrumental acquisition parameters3,4 (Box 1). These developments have resulted in a high degree of reproducibility in mass accuracy and peak intensities (Supplementary Fig. 1 online). Recently, we have optimized our protocol to be able to increase the number of molecular species analyzed by collecting two sets of sections, covering one set of sections with sinapinic acid for optimal detection of proteins and adjacent sections with 2,5-dihydroxybenzoic acid (DHB) matrix for the optimal detection of low-mass species, including peptides. Approximately 1,000 peaks can be observed in each dataset (Fig. 1). Furthermore, the sections are collected at an equal distance, 200 μm instead of 400–500 μm used previously, thus enabling the use of virtual z-stacks and three-dimensional (3D) volume renderings to investigate differential localization patterns in much smaller brain structures such as the substantia nigra and the interpeduncular nucleus. Here we present our optimized step-by-step procedure based on previous work in our laboratory2, describing how to make 3D volume reconstructions of MALDI IMS data, as applied to the rat brain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Caprioli, R.M., Farmer, T.B. & Gile, J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 69, 4751–4760 (1997).
Crecelius, A.C. et al. Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J. Am. Soc. Mass Spectrom. 16, 1093–1099 (2005).
Aerni, H.R., Cornett, D.S. & Caprioli, R.M. Automated acoustic matrix deposition for MALDI sample preparation. Anal. Chem. 78, 827–834 (2006).
Groseclose, M.R., Andersson, M., Hardesty, W.M. & Caprioli, R.M. Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J. Mass Spectrom. 42, 254–262 (2007).
Reyzer, M.L., Hsieh, Y.S., Ng, K., Korfmacher, W.A. & Caprioli, R.M. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 38, 1081–1092 (2003).
Khatib-Shahidi, S., Andersson, M., Herman, J.L., Gillespie, T.A. & Caprioli, R.M. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal. Chem. 78, 6448–6456 (2006).
Hankin, J.A., Barkley, R.M. & Murphy, R.C. Sublimation as a method of matrix application for mass spectrometry imaging. J. Am. Soc. Mass Spectrom. 18, 1646–1652 (2007).
Schwartz, S.A., Reyzer, M.L. & Caprioli, R.M. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 38, 699–708 (2003).
Chaurand, P., Norris, J.L., Cornett, D.S., Mobley, J.A. & Caprioli, R.M. New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J. Proteome Res. 5, 2889–2900 (2006).
Acknowledgements
We thank D.S. Cornett (Vanderbilt University) for custom data preprocessing software, B. Dawant (Vanderbilt University) for assistance with registration functions, P. Chaurand and L. Manier (Vanderbilt University) for assistance with MS/MS analyses, J. Morgan (St. Jude Children's Research Hospital, Memphis, Tennessee) for the kind gift of PEP-19 antibody, and J. Smith (Vanderbilt University) for assistance with using the 3D Amira software. This work was supported by The National Parkinson Foundation Center of Excellence at Vanderbilt, the National Institute of General Medical Science (GM58008-08), the Department of Defense (W81XWH-05-1-0179) and the National Cancer Institute of the US National Institutes of Health (CA86243-03).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Methods (PDF 925 kb)
Supplementary Video 1
The movie shows the co-registered 3D volumes of MS imaging data of SP and PEP-19 in the ventral midbrain rotating around its x axis. The movie starts with a frontal view of the midbrain, showing the tissue distribution of SP in green and PEP-19 in purple. Overlap of peptide and protein distribution is signified by a white appearance. (MOV 1087 kb)
Supplementary Video 2
The 3D volumes of MS imaging data of SP and PEP-19 in the ventral midbrain rotate around the y axis. The movie starts with a frontal view of the midbrain, showing the tissue distribution of SP in green and PEP-19 in purple. Overlap of peptide and protein distribution is signified by a white appearance. (MOV 961 kb)
Rights and permissions
About this article
Cite this article
Andersson, M., Groseclose, M., Deutch, A. et al. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat Methods 5, 101–108 (2008). https://doi.org/10.1038/nmeth1145
Issue Date:
DOI: https://doi.org/10.1038/nmeth1145
This article is cited by
-
Comparative lipid profiling of murine and human atherosclerotic plaques using high-resolution MALDI MSI
Pflügers Archiv - European Journal of Physiology (2022)
-
Light sheet fluorescence microscopy guided MALDI-imaging mass spectrometry of cleared tissue samples
Scientific Reports (2020)
-
Self-assembling peptides imaged by correlated liquid cell transmission electron microscopy and MALDI-imaging mass spectrometry
Nature Communications (2019)
-
MALDI MS Imaging at Acquisition Rates Exceeding 100 Pixels per Second
Journal of the American Society for Mass Spectrometry (2019)
-
Distribution of cell wall hemicelluloses in the wheat grain endosperm: a 3D perspective
Planta (2018)