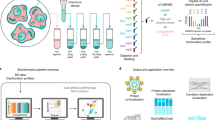
Abstract
We report a tool for the analysis of subcellular proteomics data, called MetaMass, based on the use of standardized lists of subcellular markers. We analyzed data from 11 studies using MetaMass, mapping the subcellular location of 5,970 proteins. Our analysis revealed large variations in the performance of subcellular fractionation protocols as well as systematic biases in protein annotation databases. The Excel and R versions of MetaMass should enhance transparency and reproducibility in subcellular proteomics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Christoforou, A. et al. Nat. Commun. 7, 9992 (2016).
Drissi, R., Dubois, M.L. & Boisvert, F.M. FEBS J. 280, 5626–5634 (2013).
Larance, M. & Lamond, A.I. Nat. Rev. Mol. Cell Biol. 16, 269–280 (2015).
Gatto, L. et al. Mol. Cell. Proteomics 13, 1937–1952 (2014).
Rodríguez-Piñeiro, A.M. et al. J. Proteome Res. 11, 1879–1890 (2012).
Andreyev, A.Y. et al. Mol. Cell. Proteomics 9, 388–402 (2010).
Carvalho, A.S. et al. Mol. Cell. Proteomics 13, 3294–3307 (2014).
Boisvert, F.M. et al. Mol. Cell. Proteomics 11, M111.011429 (2012).
Pinto, G. et al. J. Proteome Res. 13, 1112–1127 (2014).
Choi, S. et al. J. Proteome Res. 11, 4983–4991 (2012).
Mulvey, C.M. et al. J. Proteome Res. 12, 1436–1453 (2013).
Bileck, A., Kreutz, D., Muqaku, B., Slany, A. & Gerner, C. J. Proteome Res. 13, 5989–6000 (2014).
Thakar, K., Karaca, S., Port, S.A., Urlaub, H. & Kehlenbach, R.H. Mol. Cell. Proteomics 12, 664–678 (2013).
Larance, M. et al. Mol. Cell. Proteomics 11, M111.014407 (2012).
Adam, S.A., Marr, R.S. & Gerace, L. J. Cell Biol. 111, 807–816 (1990).
Ramsby, M. & Makowski, G. Cold Spring Harb. Protoc. 2011, prot5592 (2011).
de Hoon, M.J., Imoto, S., Nolan, J. & Miyano, S. Bioinformatics 20, 1453–1454 (2004).
Davis, J. & Goadrich, M. in Proc. 23rd International Conference on Machine Learning 233–240 (ACM, 2006).
Fawcett, T. Pattern Recognit. Lett. 27, 861–874 (2006).
de Souto, M.C., Jaskowiak, P.A. & Costa, I.G. BMC Bioinformatics 16, 64 (2015).
Acknowledgements
The authors thank J. Olweus, K.J. Malmberg and J. Trimmer for helpful discussions and critical reading. Grant support was provided by the K.G. Jebsen Foundation (F.L.-J.), Helse Sør-Øst (2013128, F.L.-J.), Norwegian Research Council (F.L.-J.), and Ministry of Education, Youth and Sports, Czech Republic (NPU I project no. LO1604, T.K. and J.S.).
Author information
Authors and Affiliations
Contributions
F.L.-J., data analysis, design of software and manuscript writing; D.d.l.R.C., data analysis, statistics and software testing; A.M., data analysis and literature review; K.S., data analysis, literature review and figure preparation; M.I., data analysis, Excel algorithms and literature review; T.K., supervision of R programming; G.A.d.S., retrieval and analysis of raw MS data; A.R.M.B., manuscript writing; K.R., statistical analysis; Q.L., Excel VBA programming; J.S., R programming, user manual for MetaMass R, data analysis and statistical analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Overlap of subcellular markers in annotation databases.
The Venn diagram shows overlap between databases in the number of proteins are listed with a single subcellular location. Lists of proteins were extracted from the annotation lookup table in Supplementary Table 2 and processed with a web-based tool to generate Venn-diagrams (http://bioinfogp.cnb.csic.es/tools/venny/,).
Supplementary Figure 2 Validation of MetaMass
The pie chart shows the similarity in the classification of dataset 1 to that reported in study 11. The dataset was processed with MetaMass using the same SVM markers used in study 1. The bar and whisker plot shows F1 scores (reported in study 1) for proteins that were mapped by both algorithms (green) and those that were mapped in study 1 but not with MetaMass (orange). The average F1 score for proteins that were mapped in both studies was 0.89 as compared to 0.75 for proteins that were mapped in study 1 only (p=3e-97, T-test two-tailed, unequal variance).
Supplementary Figure 3 HPA annotations have high numbers of nuclear markers
The pie charts show the frequency of markers for indicated subcellular locations in indicated marker set. The data were extracted from Supplementary Table 2. See also legend to Fig. 1.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Protocol (PDF 2867 kb)
Supplementary Tables 1–14
Supplementary Tables 1: Overview of methodology described in 92 articles on subcellular proteomics. Supplementary Tables 2: All Uniprot entries for the reviewed human proteome with full-text and processed annotations on their subcellular localization from Uniprot, GO and the Human Protein Atlas. The table also contains all marker panels used to generate Fig. 1 and 2. Supplementary Tables 3: All datasets included in the study. Protein identifiers and normalized MS signal values measured in 11 independent studies on subcellular proteomics. Supplementary Tables 4: Statistics output from MetaMass Excel for Fig. 1a. Supplementary Tables 5: Mapping result from MetaMass Excel for Fig. 1a. Supplementary Tables 6: Statistics output from MetaMass Excel for Fig. 2a. Supplementary Tables 7: Mapping result from MetaMass Excel for Fig. 2a. Supplementary Tables 8: Clustering results (cluster 3.0) used as input in MetaMass Excel to perform the analysis shown in Fig. 1 and 2. Supplementary Tables 9: Import/protein identifier conversion table for formatting of user-generated MS data for analysis with MetaMass (R and Excel versions, see user MetaMass user manual p7). Supplementary Tables 10: List of links and commands for installation and use of MetaMass R. The links open webpages for installation or R and R-studio. The commands can be copied directly into R-studio for installation and use of MetaMass R. Supplementary Tables 11: Template for user-generated annotation table for use in MetaMass R. (see MetaMass user manual p25). Supplementary Tables 12: Source data used to generate Fig. 1. Protein identifiers were converted from mouse to human orthologues, and MS signal values were normalized. The data can be copied directly into MetaMass Excel together with the corresponding clustering result in supplementary Table 8 to reconstruct the analysis shown in Fig. 1. Supplementary Tables 13: Source data used to generate Fig. 2a,b. The data correspond to those in supplementary Table 3, but are limited to proteins that occur two or more of datasets 2-11. The data can be copied directly into MetaMass Excel together with the corresponding clustering result in supplementary Table 8 to reconstruct the analysis shown in Fig. 2a,b. Supplementary Tables 14: Data table output from MetaMass R when the tool was used with the same markers and data used to generate the results in Fig. 2a. see MetaMass user manual p22 (supplementary Protocol). (XLSX 15724 kb)
Supplementary Software
MetaMass Excel Macro-enabled Excel workbook for automated analysis of subcellular proteomics data. (XLSM 4189 kb)
Source data
Rights and permissions
About this article
Cite this article
Lund-Johansen, F., de la Rosa Carrillo, D., Mehta, A. et al. MetaMass, a tool for meta-analysis of subcellular proteomics data. Nat Methods 13, 837–840 (2016). https://doi.org/10.1038/nmeth.3967
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3967
This article is cited by
-
Deep and fast label-free Dynamic Organellar Mapping
Nature Communications (2023)
-
SubCellBarCode: integrated workflow for robust spatial proteomics by mass spectrometry
Nature Protocols (2022)
-
Ultrastructural and proteomic profiling of mitochondria-associated endoplasmic reticulum membranes reveal aging signatures in striated muscle
Cell Death & Disease (2022)
-
Spatial-proteomics reveals phospho-signaling dynamics at subcellular resolution
Nature Communications (2021)
-
Spatial proteomics: a powerful discovery tool for cell biology
Nature Reviews Molecular Cell Biology (2019)