Abstract
Galanin is widely distributed in enteric nerve terminals lining the gastrointestinal tract1,2. We previously showed that pathogenic Escherichia coli, but not normal commensal organisms, increase galanin-1 receptor expression by epithelial cells lining the colon3 (i.e., colonocytes). When present, galanin-1 receptor activation by ligand causes colonocyte Cl− secretion 4. We herein demonstrate that disparate pathogens including Salmonella typhimurium and Shigella flexerii also increase colonocyte galanin-1 receptor expression, whose activation is responsible for a principal component of the increased colonic fluid secretion observed. Although eliminating the GAL1R gene by homologous recombination does not alter basal colonic fluid secretion, removal of one or both alleles completely attenuates the increase in fluid secretion due to infection with enteric pathogens. Galanin-1 receptor up-regulation therefore represents a novel mechanism accounting for the increased colonic fluid secretion observed in infectious diarrhea due to several different pathogens.
Similar content being viewed by others
Main
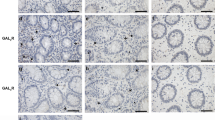
We initially determined whether other enteric pathogens known to up-regulate NF-κB, such as Salmonella typhimurium 5 and Shigella flexerii 6, increase galanin-1 receptor (Gal1-R) expression in epithelial cells lining the mouse colon similarly to what we previously observed for enterohemorrhagic E. coli (EHEC) 3. Specifically, 106 organisms in 200 μl of pH-buffered sodium phosphate were introduced into the stomachs of non-fasted mice by gavage as previously described3. We used the novel technique of quantitative immunohistochemistry (Q-IHC)7 to determine the kinetics of Gal1-R expression in murine colonocytes subsequent to pathogen infection. Whereas there is no evidence of Gal1-R expression by colonocytes derived from control C57BL/6J mice despite their being colonized by normal commensal organisms3, EHEC, Salmonella and Shigella all increased the expression of this protein (Fig. 1a). The time course of Gal1-R expression induced by each pathogen was unique. EHEC caused a rapid yet transient increase in Gal1-R expression, with peak concentrations observed 3 days post gavage, followed by a return to normal concentrations ∼3 days later. In contrast, Salmonella and Shigella infection resulted in significantly higher concentrations of Gal1-R expression, with peak concentrations observed 6 and 4 days post-gavage respectively (Fig. 1a). Although Gal1-R expression induced by Shigella decreased thereafter, that due to Salmonella remained high. For both EHEC and Shigella, infection was transient and self-limited, whereas an increasing proportion of mice died of systemic Salmonella 6 days post-gavage (data NS).
a Kinetics of Gal1-R expression by epithelial cells lining the colon after gavage with the indicated pathogen. Quantitative immunohistochemistry was performed as described in Methods, with data presented as means ± SEM for a minimum of 3 separate animals per data point in the value-less measure of energy units per pixel (eu/pix). b, Colonic fluid secretion in mice whose closed loop was injected with saline (□) or galanin (░) Inset shows representative colons from Salmonella-infected mice injected with either saline or galanin. c, Effect of concomitant parenteral administration of dexamethasone or intraluminal administration of commercially available antibody to galanin. For the middle and right panels, data represents the means±SEM of a minimum of 5 separate experiments for the amount of colonic fluid generated over a 4 h period.
To determine if increased Gal1-R expression results in increased colonic fluid secretion, we next performed closed loop experiments. Detailed studies were performed on mice infected with Salmonella because this pathogen caused the greatest amount of Gal1-R expression over the longest period of time (Fig. 1a). Overall, colonic fluid secretion increased subsequent to Salmonella gavage, with the increase (Fig. 1b) paralleling the increase in Gal1-R expression (Fig. 1a). The addition of saturating concentrations of galanin, potentiated the increase in colonic fluid production (Fig. 1b), again in approximate relation to the amount of Gal1-R present (Fig. 1a). Whereas exogenous galanin had no effect on colonic fluid production in control mice, this ligand increasingly potentiated the amount of fluid generated such that peak concentrations were observed 6 days post-Salmonella gavage (Fig. 1b). Basal concentrations of colonic fluid secretion in uninfected mice were 31 ± 2 mg/cm over a 4 h period, and were not altered by the addition of intraluminal galanin (30 ± 2 mg/cm/4 h). In contrast, basal colonic fluid secretion was ∼3-fold higher, and galanin-potentiated secretion ∼5-fold higher, 6 days post Salmonella gavage.
We have previously shown that the NF-κB-inhibitor dexamethasone 8,9,10 completely attenuates EHEC-induced activation of this transcription factor and the associated increases in Gal1-R expression in murine colonocytes3. We therefore treated mice with 0.15 μg/mg dexamethasone administered intraperitoneally every 12 hours for 6 days post-Salmonella gavage. Similar to what we have reported for EHEC, dexamethasone markedly reduced the increase in Gal1-R expression in infected mice (data not shown) such that colonic fluid secretion in the presence of exogenous galanin was reduced commensurately (50 ± 5 mg/cm/4 h in the presence of dexamethasone vs. 177 ± 9 in the absence of dexamethasone).
To confirm the contribution of Gal1-R activation to the excessive fluid secretion observed, we instilled commercially available antibody to galanin into the lumen of Salmonella-infected treated mice. Whereas colonic fluid secretion in mice 6 days after Salmonella gavage was ∼3-fold greater than observed in control mice, intraluminal administration of galanin antibody completely eliminated the Salmonella-induced increase in colonic fluid secretion (Fig. 1c). Specifically, the amount of colonic fluid generated over 4 h from the colonic loops of Salmonella-infected mice in the presence of galanin antibody (39 ± 6 mg/cm/4 h) was not significantly different from that observed in untreated control mice processed in parallel (37±4 mg/cm/4 h) (Fig. 1c).
Colonic fluid secretion was likewise determined in wild type mice at the time of maximal Gal1-R expression after gavage with EHEC and Shigella (Table 1). Overall, galanin-potentiated colonic fluid secretion was 2.2-fold higher post gavage with EHEC, and 2.9-fold higher after Shigella gavage, at the pathogen-specific point of maximal Gal1-R expression (Fig. 1a). Thus all 3 enteric pathogens share the common feature of increasing colonic epithelial cell Gal1-R expression, with the expression of this protein allowing for increased amounts of colonic fluid secretion.
Antagonists specific for the Gal1-R do not exist, with putative antagonists identified before the cloning of this receptor subtype having been found to have partial agonist activity (reviewed in ref. 4). To dissect out the specific contribution of the Gal1-R to infectious diarrhea, we eliminated one or both alleles of the GAL1R gene from C57BL/6J mice (i.e., generating GAL1R+/− or GAL1R−/− mice). As described in Methods, this was accomplished by inserting a neomycin-resistance gene 144 bp into exon 1 followed by electroporating the relevant construct into ES cells (Fig. 2a). Appropriate clones were identified, expanded, and injected into pseudopregnant mice. After confirmation of germ line transmission in the chimeric offspring, heterozygotes were back bred into C57BL/6J mice. Mice were used for study only after back breeding with C57BL/6J mice for a minimum of 20 generations. At no point was any difference in development or weight gain observed between wild type, GAL1R+/−, or GAL1R−/− mice; whereas survival in all genotypes was identical for up to 8 months, the longest time point considered (n = 5 per genotype, data NS). We previously demonstrated that Gal1-R are normally expressed by islet cells within the murine pancreas 3. Confirmation of murine genotype was confirmed in all instances after the completion of the relevant investigation by evaluating for the immunohistochemical presence of Gal1-R in pancreatic islets (Figs. 2b-d), as well as by genotyping of tail-derived DNA (data NS).
a, The relative location of the 3 exons comprising the murine GAL1R gene is shown under Native, with the relevant portion of the targeting knock-out vector shown underneath. Arrows labeled A–D identify the relative location of PCR primers used for genomic analysis of the ES cell lines after selection in aminoglycoside G-418. Representative PCR reactions with the indicated primers on ES cell genomic DNA is shown under ES cell analysis, and serves to indicate the relative frequency by which the mutant allele could be detected. Confirmation of GAL1R gene elimination was determined immunohistochemically in murine pancreatic islets (b–d) and colonic epithelial cells (e–g) 3 days after infection with EHEC. Tissues are from GAL1R+/+ (b and e), GAL1R+/− (c and f), and GAL1R−/− (d and g) mice. Gal1-R are normally expressed by pancreatic islet cells 3, with decreased presence detected in mice lacking one or both alleles for the GAL1R gene. The amount of Gal1-R up-regulation in colonic epithelia induced by EHEC infection is directly related to animal genotype (e–g). Magnification, ×1000.
Elimination of both alleles for the GAL1R gene completely eliminated Gal1-R protein expression in colonic epithelial cells in response to pathogenic infection (Table 1; Fig. 2g). In contrast some, but not all, of the nuclei and surrounding regions were immunopositive for Gal1-R expression in infected heterozygote mouse intestinal epithelial cells (Fig. 2f), a phenomenon not observed in wild type (Fig. 2e) or GAL1R−/− mice (Fig. 2g). Although the significance of this peri-nuclear expression is not clear, we determined the amount of Gal1-R present by performing quantitative immunohistochemistry (Q-IHC)7. In performing this technique we can only evaluate 100 ×100 pixel regions; because we previously showed that Gal1-R modulates Cl− secretion only when expressed at the level of the cell membrane4, we restricted Q-IHC analysis to cellular regions not overlying the nuclei. When performed in this manner, the amount of Gal1-R present in infected heterozygous mice was no different than observed in GAL1R−/− mice (Table 1). However, to determine the physiological consequence of eliminating one or both alleles of the GAL1R gene, we next performed closed loop studies and directly assessed colonic fluid secretion in animals of all 3 genotypes.
Basal colonic fluid secretion in uninfected mice was not altered as a function of GAL1R genotype (Table 1). Yet elimination of even one allele for this gene markedly attenuated the increase in pathogen-induced colonic fluid secretion (Table 1). To confirm that the colonic mucosa of genetically manipulated mice responds normally to other secretagogues, tissues were studied in an Ussing chamber. In wild-type mice, short circuit current (Isc) increased 2.1 ± 0.2-fold in response to carbachol and 3.7 ± 0.4-fold in response to forskolin. There was no difference in the kinetics or magnitude of secretagogue-induced increases in Isc in mice missing one or both alleles for the GAL1R gene. Specifically, 100 μM carbachol increased Isc 2.3 ± 0.5-fold and 2.4 ± 0.2-fold in GAL1R+/− and GAL1R−/− mice, respectfully. Similarly, 1 μM forskolin increased Isc 3.8 ± 0.4-fold and 3.7 ± 0.3-fold in GAL1R+/− and GAL1R−/− mice. No differences in responsiveness to these secretagogues were seen subsequent to infection with enteric pathogens (data NS). Yet whereas 1 μM galanin had no effect on Isc in wild-type mice, it increased Isc 5.1 ± 0.3-fold subsequent to infection with EHEC and 8.4 ± 0.7-fold in response to Salmonella infection. Similar to what we observed for colonic fluid secretion (Table 1), elimination of even 1 allele of the GAL1R gene completely eliminated the ability of galanin to increase Isc in GAL1R+/− and GAL1R−/− mice infected with these enteric pathogens (data NS).
Overall no difference in animal survival or outcome was observed between wild type and GAL1R−/− mice subsequent to infection with the enteric pathogens studied herein. Whereas infections with EHEC or Shigella were self-limited, increasing numbers of mice infected with Salmonella died of systemic infection irrespective of genotype. Salmonella infection in particular is known to induce a host colonic inflammatory response11,12. We therefore studied if eliminating the GAL1R gene altered the inflammatory response in GAL1R+/− and GAL1R−/− mice. To do this we measured colonic myeloperoxidase (MPO) concentrations, a measure of overall neutrophil presence13,14. There was no difference in basal MPO concentrations between wild type and GAL1R−/− mice exposed to their own normal commensal organisms (12.9 ± 1.1 vs. 15.5 ± 1.9 OD405 units/g tissue). After 6 days of exposure to Salmonella by gavage, MPO concentrations increased to 29.0 ± 2.7 OD405 units/g tissue in wild-type mice and 46.5 ± 12.0 OD405 units/g tissue in GAL1R−/− mice. Thus elimination of the GAL1R gene did not alter the animal's ability to mount an inflammatory response.
Thus this paper directly demonstrates that Gal1-R up-regulation by epithelial cells lining the colon is critical to mediating the bulk of the excess colonic fluid secretion observed subsequent to infection with disparate enteric pathogens. Eliminating the GAL1R gene does not alter the responsiveness of colonic epithelia to other secretagogues (such as carbachol and forskolin), and does not alter the host inflammatory response subsequent to pathogen infection. These observations suggest that the identification of Gal1-R-specific antagonists may result in new pharmacological therapies for the treatment of infectious diarrhea.
Methods
Mice and reagents.
Specific pathogen-free C57BL/6J mice used for breeding with our chimeric mice (described below) were from the Jackson Laboratory (Bar Harbor, Maine). Enterohemorrhagic Escherichia coli (EHEC, serotype 0157:H7) were provided by J. Kaper (Center for Vaccine Development, University of Maryland, Baltimore, Maryland); whereas Salmonella typhimurium and Shigella flexnerii were from ATCC (Rockville, Maryland). Immunohistochemical reagents were from DAKO (Carpinteria, California). All other reagents were from Sigma (St. Louis, Missouri). The UIC Animal Care Committee approved this study, and all care was provided in accordance with institutional guidelines.
Creation of GAL1R gene homozygous null mutations.
A primary murine genomic library containing 10.8 × 106 independent recombinants and amplified once was obtained from. H. Helmich (University of Texas Medical Branch, Galveston, Texas). This library had been made from high molecular weight DNA extracted from mouse liver (strain 129/sv) subcloned into λGem-11 (Promega, Madison, Wisconsin) at the XhoI site. We screened this library using a full-length random-primed, 32P-labeled human Gal1-R cDNA probe, originally isolated from human colon as previously described 15. Approximately 2 × 106 clones were screened at 55 °C, followed by washing in 2× SSC at 37 °C and 0.1× SSC at 55 °C. We isolated a clone containing 3 Kb of 5′ flanking region, the entire first exon, and 2 Kb of the first intron (∼6 Kb total size). Using the vector pNTK supplied to us by V. Tybulewicz (University of Cambridge, UK)16, we inserted a 954 bp fragment including 144 bp of translated exon 1, the entire 5′UTR, and 744 bp of 5′ flanking region upstream of the neomycin gene. We eliminated 84 bp of exon 1, and placed the remainder of this exon and ∼2.5 Kb of intron 1 between the Neo and TK genes (Fig. 2a). The vector was linearized and sent to Genome Systems (St. Louis, Missouri) for electroporation into ES/129 cells. After neomycin selection, 192 separate clones were provided to us for genetic analysis. We determined that 147/192 (77%) ES clones contained the neomycin gene, but only 3/192 (<2%) underwent homologous recombination and not random insertion (Fig. 2a). Genome Systems was then informed as to which clones should be used for injection into a pseudopregnant mouse (clones #111, 134, 279). Chimeric offspring were obtained from all 3 clones, and all have given birth to heterozygotes (+/−) indicating germ line transmission. Heterozygote offspring were then back bred with C57BL/6J mice for a minimum of 20 generations, and then mated as necessary to generate mice of the desired genotype.
Infection of mice by gavage.
Pathogenic bacteria were cultured until they were in mid-log growth (OD600 0.1 – 0.2) (ref. 17). As previously described, pathogens were re-suspended in 1.2 M sodium phosphate (pH 8.0), and 1 × 106 organisms in 200 μl introduced by gavage using a Teflon-tipped 10 cm long needle to a lightly anesthetized animal3. After recovery, mice were then kept in microisolator cages with free access to food and water. Access to food but not water was eliminated 24 hours before sacrifice.
Quantitative immunohistochemistry.
A standard three-stage indirect immunoperoxidase technique was used for all immunohistochemistry18. We used an anti-peptide antibody specific for the Gal1-R at a dilution of 1:500 as previously described for 60 min at 22 °C3. Chromogen quantification was performed as previously described7. Briefly, digital images of the treated tissues were acquired at 1000× using a Microlumina Ultra Resolution Scanning Digital Camera [3380 × 2700 pixels (Leaf Systems, Fort Washington, Pennsylvania)] attached to a Nikon E600 microscope system. Chromogen abundance (i.e., energy) was quantified by calculating the cumulative signal strength within 3 randomly selected 100 × 100 pixel regions located over regions not containing nuclei. The amount of chromogen per pixel was determined by subtracting the energy of the control slide (i.e., not exposed to primary antibody) from that in the homologous region of the experimental slide (i.e., exposed to primary antibody). Chromogen quantity is expressed in the value-less units of “energy units per pixel” (eu/pix).
Electrophysiology
As previously described19, full-thickness pieces of proximal colon were resected and studied in Ussing chambers. Tissues were mounted on 4-mm internal diameter rings and incubated at 37 °C in Ringer's solution that was continually gassed with 95% O2/5% CO2. Transepithelial resistance was determined as described previously3 by applying a 2-second pulse of 100 μA under current clamped conditions. Short circuit current (Isc) was measured and recorded every minute until the development of a stable baseline, at which point changes due to the addition of various secretagogues was determined under voltage-clamped conditions. Results are expressed as means ±SEM for between 7 and 11 separate animals per condition.
Evaluation of colonic fluid secretion.
After fasting for 24 h, mice were anesthetized using sodium pentobarbital (50 μg/mg) delivered in 2 divided doses as an intraperitoneal injection. The adequacy of anesthesia was assessed by inter-digital toe-pinch as described20, a small laparotomy incision was made, and the most distal and proximal segments of the colon exposed as described19. Ligatures were placed on the most distal and proximal aspects of the colon generating a loop ∼5 cm in length. A 26 gauge TB needle was inserted through the area defined by the proximally placed suture before completely tying it off. 200 μl of 1 μM galanin or saline (as control) was injected into the colonic lumen, the needle withdrawn, and suture tightened. The bowel was placed back into the peritoneal cavity and the laparotomy incision closed. Mice were sacrificed 4 h later by CO2 asphyxiation, the colonic loops removed, and their lengths and weights recorded. As previously described, fluid secretion was calculated by determining the loop weight (mg) to length (cm) ratio 19,21.
Intestinal inflammation analysis.
The intensity of intestinal inflammation was determined by measuring total tissue myeloperoxidase (MPO) content, a marker of neutrophil presence, using a modification of what has been previously published13,14. Freshly resected colon was washed in ice-cold Ringer's solution and then homogenized in PBS containing 1% Triton X-100. Samples were clarified by centrifugation at 10,000 rpm for 5 min and pH adjusted to 4.2 using 1 M sodium citrate. MPO activity was assayed by adding equal volumes of 1 mM 2,2′-azino-di-(3-ethyl) dithiazoline sulfonic acid and 10 mM H2O2 in 100 citrate (pH 4.2), and the color quantitated spectrophotometrically at 405 nm. Results are expressed in terms of the OD at 405 nm obtained per gram of homogenized tissue.
References
Hoyle, C.H. & Brunstock, G. Galanin-like immunoreactivity in enteric neurons of the human colon. J. Anat. 166, 23–33 (1989).
Bauer, F.E. et al. Distribution and molecular heterogeneity of galanin in human, pig, guinea pig, and rat gastrointestinal tracts. Gastroenterology 91, 877–883 (1986).
Hecht, G. et al. Pathogenic Escherichia coli increase Cl− secretion from intestinal epithelia by upregulating galanin-1 receptor expression. J. Clin. Invest. 104, 253–262. (1999).
Benya, R.V., Marrero, J.A., Ostrovskiy, D.A., Koutsouris, A., & Hecht, G. Human colonic epithelial cells express galanin-1 receptors which when activated cause Cl− secretion. Am. J. Physiol. 276, G64–G76 (1999).
Zunjic, M., Savkovic, S.D., Koutsouris, A., & Hecht, G. Increased IL-8 expression induced by Salmonella, as compared to pathogenic E. coli, results from synergism between NF-κB and NF-IL6. Gastroenterology 112, A1128 (1997).
Dyer, R.B., Collaco, C.R., Niesel, D.W., & Herzog, N.K. Shigella flexneri invasion of HeLa cells induces NF-κB DNA binding activity. Infect. Immun. 61, 4427–4433 (1993).
Matkowskyj, K.A., Schonfeld, D., & Benya, R.V. Quantitative immunohistochemistry by measuring cumulative signal strength using commercially available software Photoshop and Matlab. J. Histochem. Cytochem. 48, 303–311 (2000).
Sacks, M. et al. Silica-induced pulmonary inflammation in rats: activation of NF-kappa B and its suppression by dexamethasone. Biochem. Biophys. Res. Commun. 253, 181–184 (1998).
Rosen, T. et al. Chronic antagonism of nuclear factor-kappa B activity in cytotrophoblasts by dexamethasone: a potential mechanism for antiinflammatory action of glucocorticoids in human placenta. J. Clin. Endocrinol. Metab. 83, 3647–3652 (1998).
Wissink, S., van Heerde, E.C., vand der Burg, B. & van der Saag, P.T. A dual mechanism mediates repression of NF-kappa B activity by glucocorticoids. Mol. Endocrinol. 12, 355–363 (1998).
Everest, P. et al. Evaluation of Salmonella typhimurium mutants in a model of experimental gastroenteritis. Infect. Immun. 67, 2815–2821. (1999).
Peterson, J.W., Boldogh, I., Popov, V.L., Saini, S.S., & Chopra, A.K. Anti-inflammatory and antisecretory potential of histidine in Salmonella-challenged mouse small intestine. Lab. Invest. 78, 523–534. (1998).
Gallin, J.I. et al. Human neutrophil-specific granule deficiency: a model to assess the role of neutrophil-specific granules in the evolution of the inflammatory response. Blood 59, 1317–1329. (1982).
Parkos, C.A., Delp, C., Arnaout, M.A., & Madara, J.L. Neutrophil migration across a cultured intestinal epithelium. Dependence on a CD11b/CD18-mediated event and enhanced efficiency in physiological direction. J. Clin. Invest. 88, 1605–1612. (1991).
Lorimer, D.D. & Benya, R.V. Cloning and quantification of human galanin-1 receptor expression by mucosal cells lining the gastrointestinal tract. Biochem. Biophys. Res. Commun. 222, 379–385 (1996).
Tybulewicz, V.L.J., Crawford, C.E., Jackson, P.K., Bronson, R.T., & Mulligan, R.C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65, 1153–1163 (1991).
Davis, L., Kuehl, M. & Battey, J. Basic methods in molecular biology. (Appleton & Lange, Norwalk, Connecticut 1994).
Jasani, B. & Schmid, K.W. Immunocytochemistry in diagnostic histopathology. (Churchill Livingstone, Edinburgh 1993).
Marrero J.A., Matkowskyj K.A., Yung K., Hecht G. & Benya, R.V. Dextran sulfate sodium-induced murine colitis activates NF-κB and increases galanin-1 receptor expression. Am. J. Physiol. 278, G797–G810 (2000).
Waynforth, H.B. & Flecknell, P.A. Experimental and surgical technique in the rat. 1–372 (Academic Press, San Diego, California 1992).
Castagliuolo, I. et al. Neurokinin-1 (NK-1) receptor is required in Clostridium difficile-induced enteritis. J. Clin. Invest. 101, 1547–1550 (1998).
Acknowledgements
This work was supported by a Student Research Award of the American Digestive Health Foundation (to K.M.); by a Post Doctoral Fellowship Award of the Crohn's and Colitis Foundation of America (to S.S.), by National Institutes of Health (NIH) grant DK-50694, a VA Merit Review and a VA Research Enhancement Awards Program (REAP) (to G.H.); and by NIH grant DK-54777, a VA Merit Review and a VA-REAP (to R.V.B.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Matkowskyj, K., Danilkovich, A., Marrero, J. et al. Galanin-1 receptor up-regulation mediates the excess colonic fluid production caused by infection with enteric pathogens. Nat Med 6, 1048–1051 (2000). https://doi.org/10.1038/79563
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/79563
This article is cited by
-
Galanin receptor 3 attenuates inflammation and influences the gut microbiota in an experimental murine colitis model
Scientific Reports (2021)
-
Galanin is a potent modulator of cytokine and chemokine expression in human macrophages
Scientific Reports (2019)
-
The evolutionary significance of depression in Pathogen Host Defense (PATHOS-D)
Molecular Psychiatry (2013)
-
Mechanisms of infectious diarrhea
Nature Clinical Practice Gastroenterology & Hepatology (2008)
-
Towards a physiology of epithelial pathogens
Pflügers Archiv - European Journal of Physiology (2002)