Abstract
The persistence of HIV replication in infected individuals may reflect an inadequate host HIV-specific CD8 + cytotoxic T lymphocyte (CTL) response. The functional activity of HIV-specific CTLs and the ability of these effector cells to migrate in vivo to sites of infection was directly assessed by expanding autologous HIV-1 Gag-specific CD8 + CTL clones in vitro and adoptively transferring these CTLs to HIV-infected individuals. The transferred CTLs retained lytic function in vivo , accumulated adjacent to HIV-infected cells in lymph nodes and transiently reduced the levels of circulating productively infected CD4 + T cells. These results provide direct evidence that HIV-specific CTLs target sites of HIV replication and mediate antiviral activity, and indicate that the development of immunotherapeutic approaches to sustain a strong CTL response to HIV may be a useful adjunct to treatment of HIV infection.
Similar content being viewed by others
Main
In humans and in animal models, the induction and maintenance of a virus-specific CD8+ T-cell response is essential for the eradication of acute viral infections such as influenza1, RSV ( ref. 2) and LCMV (ref. 3), and for the control of persistent infections such as CMV4,7, HSV (ref. 8), and EBV (refs. 9,10). Although direct evidence for an anti-viral effect of cytotoxic T lymphocytes (CTLs) in HIV-infected individuals has yet to be demonstrated, inferential evidence suggests that CD8+ CTLs exert substantial anti-HIV activity. After primary infection, the appearance of HIV-specific CD8+ CTLs coincides with a reduction in viremia and an increase in CD4+ T-cell numbers, and occurs before HIV-specific neutralizing antibodies can be detected11,12,13,14. Infected individuals with a prolonged phase of asymptomatic infection, called long-term non-progressors, maintain strong HIV-specific CTL responses15,16,17, whereas individuals with progressive disease lose detectable CTL responses or harbor HIV variants with 'escape' mutations in the epitopes recognized by their CTLs17,18,19,20,21. CTLs can be detected in lymphoid organs, which are the chief sites of HIV replication22. Moreover, plasma RNA viral load, an indicator of progressing disease, correlates inversely with the magnitude of HIV-specific CD8+ CTL activity14,23.
Several observations have suggested that CD8+ CTLs have a less important role in limiting progression of HIV infection. Although primary infection is associated with a vigorous CTL response, the HIV-specific CTLs seem to be preferentially localized in the blood rather than lymph nodes, which may facilitate establishing and maintaining a reservoir of productively infected cells at relatively privileged sites within lymph nodes24. HIV Nef reduces cell surface class I MHC expression, which can interfere with the recognition and elimination of infected cells by CTLs ( ref. 25). Some investigators believe that HIV-specific CD8+ CTLs not only are irrelevant to viral control, but also may be deleterious to the host by contributing to depletion of CD4+ T cells or by damaging bystander cells in the lung or central nervous system26,27,28. Thus, the role of CTLs in HIV infection and the potential limitations of this CTL response in containing virus require further analysis.
Adoptive transfer of antigen-specific T cells can reconstitute immune responses to human pathogens such as CMV and EBV, and can be used to directly assess the contribution of specific T-cell populations to host defense5,29,30. Thus, we transferred HIV-specific CD8+ CTLs to determine if CTLs can 'home' to sites of viral replication and mediate anti-viral activity in HIV-infected individuals31,32. An initial study to assess safety was done by administering escalating doses of autologous Gag-specific CTL clones genetically modified to express hygromycin phosphotransferase and herpes simplex thymidine kinase, making it possible to ablate the cells in vivo with ganciclovir if toxicity was observed. Unfortunately, the in vivo activity of the transferred CTLs could not be assessed, as persistence was limited by the induction, after three or four cell infusions, of a host CTL response to the foreign proteins hygromycin phosphotransferase and herpes simplex thymidine kinase32. Therefore, we administered here three infusions of unmodified autologous HIV Gag-specific CD8+ CTL clones in escalating doses to determine safety, followed by two infusions of CD8+ CTLs genetically modified (using the LN retrovirus) to express the neomycin phosphotransferase (neo) gene, to assess transfer of immunity, persistence in the peripheral blood, and migration to lymph nodes.
Clinical protocol for adoptive transfer of CD8 + CTLs
HIV Gag-specific CD8+ CTL clones were isolated from three HIV seropositive individuals with CD4+ cell counts between 100 cells/mm3 and 500 cells/mm3. Each subject was on a stable regimen of anti-retroviral drug therapy and showed no evidence of opportunistic infection. All three lacked demonstrable HIV-specific T-helper cell responses (Fig. 1a). The CTL clones were MHC class I restricted, and recognized epitopes in either the p24 or p17 subunit of HIV Gag (Table). Three infusions of unmodified CTL clones were administered at 2-week intervals in doses of 3.3×108 cells/m2, 1×109 cells/m2, and 3.3×109 cells/m2 (per meter squared of body surface area). One week later, two infusions of neo-modified CTLs were given with a 1-week interval, in doses of 1×109 cells/m2 and 3.3×109 cells/m2, and a lymph node biopsy was done 4 days after the final infusion. The CTL infusions were well tolerated, although all patients experienced symptoms of fever, myalgias and/or arthralgias, lasting less than 48 hours, after some of the infusions, as previously observed in recipients of HIV-specific CTLs. No substantial changes in CD4+ T-cell counts were noted during or after CTL immunotherapy (Fig. 1b).
a, Analysis of HIV-specific T helper-cell responses. Patient PBMC were assayed for proliferative responses to HIV-1 p24 and gp160 antigens, PHA and anti-CD3 monoclonal antibody stimulation. A stimulation index of greater than 5 indicates a positive response37. b, CD4+ T-lymphocyte counts in peripheral blood before, during and after the T-cell infusions. ▪, patient 1; ●, patient 2; ▴, patient 3.
Transfer of CD8 + CTLs augments cytolytic activity
To determine if the adoptive transfer of CTLs augmented circulating HIV-specific cytolytic activity, peripheral blood mononuclear cells (PBMC) were obtained 1 day before and 1, 6 and 14 days after infusion of the highest T-cell dose (3.3×109 cells/m2), and were assayed for lytic activity against autologous EBV-transformed lymphoblastoid cell lines (LCL) expressing either HIV Gag or an irrelevant CMV protein ( Fig. 2a). Although HIV-specific cytolytic activity was not detectable by direct assay of the PBMC before therapy, lytic activity was evident if the PBMC sample before therapy was 'spiked' to contain 0.3% of the HIV Gag-specific CTL clone subsequently used in therapy. PBMC demonstrated direct HIV-specific cytolytic activity at 1 and 6 days after T-cell infusion. However, by day 14, lytic activity returned almost to the baseline levels (before therapy). The actual frequency of neo-modified CD8+ T cells in the peripheral blood samples obtained after CTL transfer was determined by two-color flow cytometry of fixed PBMC using a phycoerythrin-labeled monclonal antibody against CD8, and PCR amplification of the neo gene followed by in situ hybridization (ISH) with fluorescein-labeled neo-specific oligonucleotides (Fig. 2c). One day after infusion of the highest dose of neo-modified CTLs (3.3×109 cells/m2), neo+ T cells constituted from 2% to 3.5% of CD8+ T cells in the peripheral blood ( Fig. 3a). Consistent with the analysis of Gag-specific cytolytic activity in the PBMC after T-cell transfer (Fig. 2a), the frequency of neo-modified CD8+ T cells in the PBMC declined each day after the infusion and were no longer detected by flow cytometry 2–3 weeks after the final T-cell infusion. Although PCR of DNA extracted from PBMC continued to detect neo-modified cells in the blood for 3 to 4 weeks after infusion, the frequencies were less than 5 cells per 106 PBMC (data not shown).
a, Evaluation of HIV-specific cytolytic activity of PBMC after infusion of the highest T-cell dose in a representative patient. PBMC collected immediately before therapy and at days 1, 7, and 14 after cell transfer were tested directly as effectors in a chromium release assay. Before the assay, an aliquot of the pre-therapy PBMC sample was also 'spiked' with 0.3% of the HIV-specific CTL clone subsequently infused into the patient. Target cells were autologous LCL infected with vac/gag (black bars) or with a vaccinia recombinant expressing an irrelevant CMV protein (vac/CMV, hatched bars). Data are shown at a PBMC:target ratio of 100:1. b, Retention of HIV Gag-specific lytic activity in neo-modified CD8+ CTLs rescued from lymph node 4 days after the last T-cell infusion. Cryopreserved LNMC from patients 2 and 3 were thawed, stimulated with anti-CD3 monoclonal antibody, and then cultured with IL-2 (50 U/ml) and G418 (1 mg/ml) for 12 days. The G418-resistant T cells were then tested against autologous LCL targets infected with vac/gag (black bars) or vac/CMV (hatched bars). Data are shown at an effector:target ratio of 4:1. c and d, Representative two-color dot-plots from patient 1 showing the frequency of neo-modified CD8+ T cells in PBMC ( c) and LNMC (d) obtained at the times indicated (upper left corners) relative to the infusion of neo-modified CTLs, and assayed by in situ hybridization for neo after PCR and labeling for CD8. The pre-treatment lymph node biopsy was obtained from the patient one year previously (d, far left). T cells positive for both CD8 and neo were counted as the percentage of gated cells and appear in the upper right quadrant of the plots. As a negative control, cells were subjected to PCR without Taq polymerase and then hybridized with target-specific probes (far right plots).
a, Temporal relationship between the percentage of CD8+ T cells marked with neo and the percentage of CD4+ T cells expressing HIV Gag mRNA in PBMC of the three patients collected before, during and after adoptive transfer of neo-modified CTL clones. Upward arrows (horizontal axis) indicate the days CTLs were infused. *, Day of lymph node biopsy. b, Representative two-color dot-plots from patient 3 showing the frequency of CD4+ T cells productively infected with HIV (expressing HIV genes) in PBMC immediately before (i and ii) and 2 days after (iii and iv) the second infusion of neo-modified CTLs, by in situ hybridization for HIV mRNA and labeling for CD4. Lymphocytes from PBMC staining positive for CD4 were gated (i and iii), and the percent of double-positive CD4+ HIV mRNA+ T cells is indicated in the upper right quadrant (ii and iv). c, Laser confocal imaging of CD4+ lymphocytes expressing HIV gag-pol mRNA obtained from an HIV-infected individual. CD4+, HIV Gag-pol-positive lymphocytes (left) emit bright, globular green cytoplasmic fluorescence, whereas the CD4+, HIV Gag-pol-negative cells (right) emit only background green fluorescence.
Transferred HIV-specific CTLs migrate to lymph nodes
Migration of the transferred CTLs to lymph nodes was assessed using two-color flow cytometry to detect neo+ CD8+ T cells in mononuclear cell suspensions (LNMC) prepared from the lymph node biopsy specimen obtained 4 days after the last T-cell infusion ( Fig. 2d). In patients 1, 2 and 3, 5%, 2.2% and 7.9%, respectively, of CD8+ T cells in LNMC were positive for neo DNA, whereas concurrent analysis of PBMC showed that only 0.5%, 0.5% and 0.7%, respectively, of CD8+ T cells were neo+ ( Fig. 3a). The analyses were done in triplicate for each timepoint, and the standard error never exceeded 0.4%. Thus, neo-modified CD8+ CTLs were present in the lymph node in excess of neo+ cells in the peripheral blood, demonstrating that the transferred cells effectively migrated to and preferentially accumulated within lymph nodes. To determine if the transferred CTLs retained function in the lymph node, LNMC suspensions from patients 2 and 3 were stimulated in vitro with anti-CD3 monoclonal antibody and cultured in G418 to select for the infused neo-modified CTLs. The resulting G418-resistant cells specifically lysed autologous LCL expressing HIV Gag (Fig. 2b).
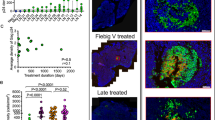
Despite migration of the transferred CTLs to lymph nodes, CTL antiviral activity might still be limited if target cells replicating HIV resided in relatively privileged sites within the lymph node architecture. Therefore, the distributions of both the CTLs and the productively infected cells within the lymph node were examined. Serial histologic sections were evaluated by in situ PCR with neo primers followed by hybridization with digoxigenin-labeled, neo-specific probes to locate the transferred CTLs, and direct ISH with digoxigenin-labeled HIV-specific riboprobes for HIV RNA to locate HIV-infected cells. The latter technique requires more than 20 copies of viral RNA per cell to detect a positive signal; thus, it detects cells infected with HIV actively expressing viral genes but does not detect latently infected cells. Aggregates of neo-modified cells and cells productively infected with HIV were found almost exclusively in the parafollicular regions of the lymph node, and not in the follicular dendritic network, which contained free virions, as shown by immunohistochemical staining for Gag p24 and by in situ hybridization for HIV RNA using 35S-labeled riboprobe (Fig. 4a–g, j). Moreover, analysis of serial histologic sections showed that the neo-modified cells were not randomly distributed but instead were selectively localized at the sites containing productively-infected cells (Fig. 4a–d). This pattern was seen with in the lymph nodes of all three patients.
Serial sections of inguinal lymph nodes collected 4 days after the second infusion of neo-modified HIV Gag-specific CTLs were analyzed. a , c and h, In situ hybridization with DIG-labeled HIV-specific riboprobes followed by staining with anti-DIG-AP to detect cells expressing HIV Gag mRNA. a, patient 2; c and h, patient 3. Original magnifications: a, ×100; c, ×40; h, ×500. b, d and i, In situ hybridization after PCR for neo using DIG-labeled oligonucleotide probes specific for the neo gene product followed by staining with anti-DIG-AP. b, patient 2; d and i, patient 3. Original magnifications: b, ×100; d, ×40; i, ×500. e, f and g, Immunohistochemistry for detection of CD4 (e) CD8 (f) and HIV p24 (g) using monclonal antobodies against human CD4, human CD8 and HIV-1 Gag p24, respectively, and staining with avidin–biotin–horseradish peroxidase complex with diaminobenzidine as the chromogen. Original magnification, ×100. j, In situ hybridization for HIV RNA using 35S-labeled riboprobe (done independently by Molecular Histology Labs, Gaithersburg, Maryland49).
Antiviral activity of transferred HIV-specific CD8 + CTLs
The transferred T cells, which demonstrated both cytolytic activity and 'homing' to sites of active infection, would be expected to have anti-viral activity. We anticipated that the anti-viral activity of the transferred CTLs might be most directly demonstrated by analyzing changes in the number of CD4+ T cells expressing HIV genes, as these productively infected cells are immediate targets for lysis by CTLs. Therefore, CD4+ T cells expressing HIV genes in samples of PBMC were stained using ISH with fluorescein-labeled HIV-specific oligonucleotides, and then quantitated by flow cytometry, or directly visualized by laser confocal microscopy (Fig. 3c). Immediately before the first infusion of neo-modified CTLs, 2.6%, 0.5% and 2.4% of the CD4+ T cells in PBMC in patients 1, 2 and 3, respectively, were productively infected with HIV (Fig. 4a). This was similar to percentages observed in 40 other asymptomatic HIV-infected individuals using this technique (mean = 0.8%; range, 0.1–3.4%; B.K.P., unpublished data). After each of the two infusions of neo-modified CTLs, the percentage of productively infected CD4+ T cells declined precipitously over the ensuing 3–4 days (Fig. 4a). A similar decline in productively infected CD4+ T cells was seen after the infusions of unmodified CTLs (data not shown). However, the frequency of transferred CTLs decreased over time, and the percentage of productively infected CD4+ T cells rebounded to baseline levels by 7 days after each infusion. By Spearman rank correlation coefficient two-tailed analysis, the frequency of neo-modified CTLs correlated inversely with the frequency of HIV-infected cells in the peripheral blood after CTL transfer (R = –0.73; P = 0.01).
Plasma HIV levels, as measured by both reverse transcriptase polymerase chain reaction (RT–PCR) and branched chain DNA (bDNA) assays, did not change substantially during the brief course of immunotherapy ( Fig. 5). This observation probably reflected several factors. Virus may have been released from target cells into the extracellular space as the transferred CTLs destroyed HIV-infected T cells, and viral production by neighboring infected cells may have transiently increased because of the cytokines (such as TNF) released by the CTLs. Indeed, transient increases in HIV bDNA were observed 1–2 days after CTL infusion (Fig. 5). Because the persistence of transferred CTLs was short-lived, replenishment of the infected pool of CD4+ T cells may have occurred before reductions in plasma virus could be observed, indicating that more sustained CTL responses may be necessary to lower plasma HIV. The production of virus by cells harboring viral variants containing mutations in sequences encoding CTL epitopes also might contribute to the sustained level of plasma virus. However, analysis of the sequence of the predominant plasma viral isolates, which were sensitive to the CTLs administered at the start of therapy, did not show 'escape' mutations during and after therapy (D.A.L., manuscript in preparation).
Discussion
Several studies have provided evidence supporting a role for HIV-specific CD8+ CTLs in controlling virus replication and preventing HIV disease progression15,16,17. However, the host response usually fails to completely contain HIV infection, indicating deficiencies in the CTL response17,18 and/or viral escape19,20,21,25. Our work here shows that adoptively transferred HIV Gag-specific CD8+ CTLs augment the endogenous CTL response, migrate to sites of viral replication in lymph nodes and result in a decline in HIV-infected CD4+ T cells in the peripheral blood, providing direct demonstrations that CTLs mediate anti-HIV activity and that the endogenous host CTL response maintained in chronic HIV infection is inadequate to cope with the reservoir of virus-infected cells established in the host.
Lymphoid tissues have been demonstrated to contain large amounts of trapped virus particles and serve as a chief site for ongoing HIV replication in CD4+ T cells and macrophages33. In individual white pulps from spleens of HIV-infected individuals, virus replication was observed in discrete foci22, and it is presumed that this also occurs in lymph nodes. Our findings demonstrate that adoptively transferred HIV-specific CTLs efficiently 'home' to lymph nodes and preferentially accumulate within the nodal architecture adjacent to cells actively replicating HIV. Evaluation of the migration of a neo-modified CD8+ CTL clone of irrelevant specificity would be needed to conclusively establish the specificity of the preferential migration. However, our data show that focal reservoirs of infection in lymphoid organs are clearly not privileged from detection by HIV-specific CTLs, indicating this is not the chief obstacle to CTL efficacy.
The decline of HIV-infected CD4+ T cells, often to undetectable levels, observed here in association with adoptive transfer of CD8+ CTLs was transient. The resurgence of HIV-infected CD4+ T cells correlated with the disappearance of transferred, neo-marked CTLs from the blood, indicating that the transient nature of the anti-viral effect reflects the limited in vivo persistence of the transferred CTLs. The disappearance of the infused CTLs from the blood probably did not result from an immune response directed against the gene-modified cells, as assays for a CTL response to the neo gene product failed to detect responses in these patients up to 28 days after the second infusion, although neo-specific CTLs were detectable in two of the three patients 42–56 days after the second infusion (S.R.R., unpublished data). This is consistent with the prolonged time required to develop immune responses to transgene products reported in other studies32,34. Moreover, the decline and subsequent rebound of HIV-infected cells associated with CTL transfer was observed after infusions of unmodified as well as neo-modified CTLs (data not shown). Although the decline in transferred CTLs in peripheral blood in part reflects migration to lymph node sites, the loss of antiviral activity over time indicates the CTLs must die or be rendered dysfunctional at these local sites of HIV replication. Obtaining serial lymph node biopsies from multiple sites to minimize sampling bias, might permit analysis of the fate of transferred cells, but this was not possible due to the morbidity of this procedure. However, studies of the adoptive transfer of CMV-specific CD8+ T-cell clones to immunodeficient bone marrow transplant patients demonstrated that transferred CTLs declined in the subset of recipients who were deficient in CMV-specific CD4+ T helper-cell responses5,30 indicating that the lack of HIV-specific CD4+ T helper-cell responses is a more likely explanation for the brief persistence of the transferred HIV-specific CTLs. Moreover, expansion of CD8+ CTLs in the presence of antigen in vitro requires supplementation with IL-2 and, in animal models, IL-2 or CD4+ T cells are required for persistence and optimal anti-viral activity of virus-specific CD8+ CTLs in vivo35,36. Long-term non-progressors who maintain vigorous CTL responses also retain HIV-specific CD4+ T helper-cell responses, whereas individuals with progressive infection, such as the patients in our study, quickly lose CD4+ T helper-cell responses37. Thus, virus-specific CD4+ T-cell responses may be essential for CD8+ CTL-mediated immunological control of HIV infection. Therefore, strategies to provide help to transferred CD8+ CTLs, such as concomitant IL-2 infusions, concurrent adoptive transfer of HIV-specific CD4+ T-helper clones genetically modified to resist HIV infection38, or genetic modification of the CD8+ CTLs to function in a CD4-deficient environment39 should improve CTL survival and elucidate the therapeutic potential of CTLs for controlling progression of HIV infection.
Methods
Clinical protocol and patient characteristics.
Protocol 827 was approved by the Fred Hutchinson Cancer Research Center Institutional Review Board, the Food and Drug Administration, and the Recombinant DNA Advisory Committee. Patients 1, 2 and 3 correspond to UPIN 11553, 11552 and 11545, respectively. Patient entry criteria were: HIV seropositive by western blot, no prior history of opportunistic infections, CD4+ T-cell count between 200 and 500 cells/mm3 at the time T-cell cultures were initiated, and a stable regimen of antiretroviral therapy for at least 4 weeks before study entry. Plasma HIV RNA was determined by HIV RNA branched chain assay (sensitivity, ≥ 500 copies; Chiron).
Characterization of HIV Gag-specific CD8+ CTL clones.
HIV Gag-specific CD8+ CTL clones were isolated and characterized as described32. The Ag specificity and MHC class I restriction of the CTL clones was determined in a standard 5-hour chromium release assay using as target cells autologous and allogeneic class I MHC-mismatched LCL that were either mock-infected, infected with a vaccinia recombinant virus expressing HIV Gag or infected with a control vaccinia recombinant virus expressing a CMV protein. Samples were assayed at an effector to target ratio of 2.5:1. Epitope mapping was done as described (Lewinsohn, D.A.L., manuscript in preparation).
Isolation and expansion of CD8+ Gag-specific CTL clones.
Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll Hypaque density gradient centrifugation, washed twice in phosphate buffered saline and resuspended in RPMI containing 25 mM Hepes, 4 mM L glutamine, and 11% human AB serum. The PBMC were cultured for 2 hours in 12-well tissue culture plates to separate adherent and nonadherent populations. The adherent cells were infected with a vaccinia recombinant virus encoding HIV Gag (vac/gag) at an MOI of 5 for 12 hours, and then exposed to a germicidal lamp to inactivate residual vaccinia virus. The nonadherent population containing mostly T cells was added to the vac/gag-infected stimulator cells, and these cultures were incubated for 7 days. Interleukin 2 (2 U/ml) was added on the third day after stimulation. The cultures were re-stimulated after 7 days with fresh ultraviolet-inactivated vac/gag-infected adherent cells, and IL-2 (5 U/ml) was added 2 and 4 days after re-stimulation. To obtain unmodified gag-specific CTL clones, an aliquot of the culture was taken 7 days after re-stimulation and plated at limiting dilution (0.5 cells/well) in 96-well round-bottomed plates containing 7.5×104 gamma-irradiated allogeneic PBMC and 1×104 gamma-irradiated allogeneic EBV-transformed B cells (LCL) derived from normal volunteer blood donors, anti-CD3 monoclonal antibody (OKT3), and 50 U/ml IL-2. To obtain neo-modified CTL clones, the remaining aliquot of the culture was re-stimulated with OKT3 and gamma-irradiated allogeneic feeder cells and exposed, on the second and third days after stimulation, to LN retrovirus supernatant produced by PA317 packaging cells (provided by Targeted Genetics, Seattle, Washington). On the fourth day, the cells were cultured in G418 (1 mg/ml) to select transduced cells. The LN-transduced cells were then plated at limiting dilution (0.5 cells/well) as described above for unmodified CTLs. After 14 days, aliquots of cloning wells positive for growth were tested for specificity for autologous vac/gag infected target cells, and T cells from those wells with gag-specific reactivity were expanded in 25-cm2 tissue culture flasks using OKT3 to provide T-cell receptor stimulation and gamma-irradiated allogeneic PBMC and LCL as feeder cells. Cultures were 'fed' on the first, fourth, seventh and tenth days with IL-2 (25–50 U/ml) and were split to maintain a density of less than 2×106 cells/ml. Sufficient cells from an individual clone were obtained after this expansion for quality control testing and to cryopreserve a cell bank that could be thawed subsequently for further expansion and reinfusion. Quality control testing included phenotype analysis to ensure the cells were CD3+CD8+CD4–; sterility assays; cytotoxicity assays to confirm class I MHC restricted gag-specific killing; and PCR of cellular DNA to ensure the clones were negative for HIV and vaccinia. LN-modified CTL clones were also analyzed by Southern blot to show integration of an unrearranged copy of the LN provirus, and were assayed and shown to be negative for replication-competent retrovirus. To expand cells for therapy, aliquots of the cell bank were stimulated in multiple 75-cm2 tissue culture flasks with OKT3 and allogeneic PBMC and LCL, and were 'fed' on the first, fourth, seventh and tenth days of a14-day stimulation cycle.
Lymphoproliferative assay.
PBMC (2×105) from patients were plated in triplicate in 96-well plates either in media alone or in presence of baculovirus recombinant gp160 (10 μg/ml, MicroGene Systems, Meridien, Connecticut), baculovirus recombinant p24 (10 μg/ml, MicroGene Systems, Meridien, Connecticut), baculovirus control antigen (10 μg/ml, MicroGene Systems, Meridien, Connecticut), phytohemagglutanin (PHA, 3 μg/ml), or anti-CD3 monoclonal antibody (30 ng/ml) in a total volume of 200 μl. After 4 days of incubation at 37 °C, cultures were 'pulsed' with 3H-thymidine (2.5 μCi/well) 18 hours before cells were collected. The stimulation index was calculated as the ratio of the mean counts per minute (cpm) of wells containing antigen to the mean cpm of wells with media alone or baculovirus control antigen.
Quantitation of neo-marked cells in blood and lymph nodes.
PBMC and LNMC (2×107 cells/ml) were incubated with biotinylated anti-CD8 (20 μg/ml; Coulter-Immunotech, Westbrook, Maine) for 30 min at 25 °C in PBS. The cells were washed twice with PBS and fixed and permeabilized in Permeafix (50 μl; Ortho Diagnostics, Raritan, New Jersey) for 60 min at 25 °C. The cells were then washed in PBS, and resuspended in 190 μl of a PCR mixture consisting of 1X PCR buffer (50 mM KCl, 10 mM Tris HCl, pH 8.3); 3.5 mM MgCl2; 0.25 mM (each) dATP, dCTP, dGTP, and dTTP; 100 pmol (each) of forward neo-specific primer (neopr.5 i: 5'–GGTGCCCTGAATGAACTGCAGGACGAGGCAGCG–3') and reverse neo-specific primer (neopr.3i: 5'–GGTCACGACGAGATCCTCG CCGTCGGGCATGCGCGCCTT–3'); and 1.0 μl (5U) of Taq polymerase (AmpliTaq; Perkin-Elmer Cetus, Norwalk, Connecticut). The samples were amplified by 25 cycles of denaturation (94 °C, 45 s), primer annealing (58 °C, 2 min) and primer extension (74 °C, 2 min). The amplification product was 399 bp in length (Genbank accession no. M28245; nucleotide positions 1818–2216 of the vector). After in vitro amplification, the cells were resuspended in 25 μl of 1X PCR buffer containing 10 μg sonicated herring sperm DNA and 100 ng neo-specific oligonucleotide probe labeled with 5'- and 3'-5-carboxyfluorescein (5'–FAM-AGCATCAGGGGCTCGCG-FAM–3'; nucleotides 2137–2153; Research Genetics, Huntsville, Alabama). HLA-DQα-specific primers GH26 and GH27 were used as control primers40. The product DNA was then denatured at 95 °C for 3 min and hybridized to the probe at 56 °C for 2 h. After hybridization, the cells were washed in a series of high-stringency buffers41 and then incubated in 20 μl of streptavidin–phycoerythrin (PE, 200 μg/ml) in PBS for 30 min at 25 °C. The cells were then filtered through a nylon mesh 37 μm in pore size and were analyzed by flow cytometry, using a Coulter XL flow cytometer. Laser excitation was 15 mW at 488 nm, and the fluorescein isothiocyanate and PE fluorescence were detected with a standard optical filter setup (550 dichroic, 525 bandpass for fluorescein isothiocyanate, and 585 bandpass for PE). At least 5,000 events were counted for each sample. Positive and negative controls were neo-modified and unmodified CD8+ CTL clones, respectively.
ISH for HIV with quantitation by flow cytometry.
CD4+ lymphocytes productively infected with HIV were quantified using ISH to HIVgag-pol RNA with analysis by flow cytometry as described42,43. PBMC were labeled with PE-conjugated anti-CD4 monoclonal antibody (PharMingen, San Diego, California) and then fixed and permeabilized with Permeafix. The cells were then hybridized with a 'cocktail' of 134 fluorescein-labeled oligonucleotides that spanned the open reading frames of HIV gag- pol, and were analyzed by two-color flow cytometry. By evaluating MT-2 cells infected with HIV at multiple timepoints after TNFα stimulation44, the limit of detection of HIV mRNA was determined to be 10 copies/cell.
Confocal microscopy of cells expressing HIV gag-pol mRNA.
After magnetic bead separation of CD4+ lymphocytes from PBMC (ref. 45), the cells were reacted with fluorescein-conjugated HIVgag-pol oligonucleotide probes42,43. HIV mRNA-positive and -negative cells were sorted by flow cytometry. The separated cells were allowed to adhere to siliconized slides and coverslips were applied. Slides were visualized with an ACAS 570 laser confocal microscope at 488 nm excitation (magnification, ×1000).
PCR followed by ISH for neo in tissue sections.
A solution containing 1× PCR buffer, 4 mM MgCl2, 0.01% gelatin, 200 μM dNTPs, 50 pM of neo-specific primers and Taq polymerase (0.15 U/μl) was prepared. A portion (30–50 μl) of this PCR mixture (at 70 °C) was applied to deparaffinized and proteinase K-treated (20 μg/ml for 20–40 min) tissue sections as described46. Coverslips were applied to the slides and surrounded with mineral oil and the slides were placed directly on the aluminum block of the thermocycler (Omnigene, Hybaid, Woodbridge, New Jersey). After 25 cycles of denaturation (94 °C, 1 min), primer annealing (55 °C, 2 min) and primer extension (72 °C, 2 min), the slides were treated for 5 min with xylenes to remove mineral oil, then for 5 min in 100% ethanol and then air-dried. The PCR product was detected by in situ hybridization using a 'cocktail' of three neo-specific oligonucleotides labeled with digoxigenin-11-dUTP (DIG; Boehringer), which were in sense orientation and internal to the PCR primer binding sequences (neo7i: 5'–GCTGTGCTCGACGTTGT–3'; neo8i: 5'–GATGCAATGCGGCGGCT–3'; and neo9i: 5'–AGCATCAGGGGCTCGCG–3'). The oligonucleotide probes (30 ng in 5× SSC, 50% formamide, 0.5% Tween-20, 100 μg/ml sonicated salmon sperm DNA and 5× Denhardt's solution) were hybridized to tissues for 4 h at 42 °C in a humidified chamber. The slides were then washed in 2× SSC, 0.5% Tween-20 for 30 min at 42 °C followed by 0.2× SSC, 0.5% Tween-20 for 20 min at 25 °C. Hybridized probe was detected with AP-conjugated anti-DIG monclonal antibodies (1,500 mU/ml) and nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate toluidinium substrate, as described44. The presence of neo DNA was indicated by a purple, cell-associated precipitate, and was visualized by incident light microscopy. Positive controls consisted of neo+ cells immobilized in paraffin wax. Negative controls for PCR included tissue processed without Taq polymerase or without neo-specific primer pairs. Identical procedures done on lymph nodes from individuals who had not received neo-modified cells also served as a negative control. ISH controls included an irrelevant oligonucleotide probe to an unrelated virus47.
ISH for cellular HIV in tissue sections.
To prepare DIG-labeled HIV-specific riboprobes for ISH, cDNAs (sense and antisense) encoding HIV gag p24 (~0.8 kb) were cloned into transcription vectors under the control of T7 and SP6 RNA polymerase promoters (pGEM; Promega, Madison, Wisconsin) using methods described47. These constructs were linearized and used as templates for in vitro transcription, in the presence of DIG-UTP. Using MT-2 cells infected with HIV at multiple timepoints44, the lower limit of detection of HIV mRNA was determined to be 20 copies/cell. Serial sections of lymph node were deparaffinized, rehydrated in Tris-buffered saline (TBS: 0.1 M Tris, pH 7.5, 0.1 M NaCl), digested with proteinase K (20 μg/ml, 37 °C, 30 min; Sigma), succinylated (5 g/l succinic anhydride in 0.1 M triethanolamine, pH 8.0), and washed in DEPC-treated water. This minimal concentration of proteinase K and digestion time permitted detection only of intracellular viral RNA, and not of antibody-complexed virus trapped in germinal centers. The remainder of the procedure was as described44. Lymph nodes from HIV-seronegative age-matched males were used as a negative control.
Immunohistochemistry.
Immunohistochemistry was done using standard procedure45,48, which included an antigen retrieval step (steaming in 1 mM citrate buffer, pH 6.0, for 10–20 min) and used monclonal antibodies (10–20 μg/ml) against CD4 (Vector Laboratories, Burlingame, California), CD8 (Dako, Carpinteria, California) and HIV-1 gag p24 (Dako, Carpinteria, California). The primary antibody was detected with isotype-specific HPO-conjugated secondary antibody and an ABC peroxidase technique using 3,3' diaminobenzidine (Vector Laboratories, Burlingame, California) as the chromagen. To control for nonspecific binding, staining was done with isotype-matched primary antibodies specific for irrelevant antigens.
References
Ada, G.L. & Jones, P.D. The immune response to influenza virus. Cur. Top. Microbiol. Immunol. 128, 1–54 (1986).
Cannon, M.J., Openshaw, P.J. & Askonas, B.A. Cytotoxic T cells clear virus but augment lung pathology in mice infected with respiratory syncytial virus. J. Exp. Med. 168, 1163–1168 ( 1988).
Byrne, J.A. & Oldstone, M.B. Biology of cloned cytotoxic T lymphocytes specific for lymphocytic choriomeningitis virus. VI. Migration and activity in vivo in acute and persistent infection. J. Immunol. 136, 698–704 (1986).
Riddell, S.R. & Greenberg, P.D. Principles for adoptive T cell therapy of human viral diseases. Annu. Rev. Immunol. 13, 545–586 (1995).
Walter, E.A. et al. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N. Engl. J. Med. 333, 1038– 1044 (1995).
Li, C.R., Greenberg, P.D., Gilbert, M.J., Goodrich, J.M. & Riddell, S.R. Recovery of HLA-restricted cytomegalovirus (CMV)-specific T-cell responses after allogeneic bone marrow transplant: correlation with CMV disease and effect of ganciclovir prophylaxis. Blood 83, 1971–1979 (1994).
Reusser, P., Riddell, S.R., Meyers, J.D. & Greenberg, P.D. Cytotoxic T-lymphocyte response to cytomegalovirus after human allogeneic bone marrow transplantation: pattern of recovery and correlation with cytomegalovirus infection and disease. Blood 78, 1373– 1380 (1991).
Posavad, C.M., Koelle, D.M., Shaughnessy, M.F. & Corey, L. Severe genital herpes infections in HIV-infected individuals with impaired herpes simplex virus-specific CD8+ cytotoxic T lymphocyte responses. Proc. Natl. Acad. Sci. USA 94, 10289– 10294 (1997).
Heslop, H.E., Brenner, M.K. & Rooney, C.M. Donor T cells to treat EBV-associated lymphoma. N. Engl. J. Med. 331, 679–680 (1994).
Papadopoulos, E.B. et al. Infusions of donor leukocytes to treat Epstein-Barr virus-associated lymphoproliferative disorders after allogeneic bone marrow transplantation. N. Engl. J. Med. 330, 1185– 1191 (1994).
Safrit, J.T. & Koup, R.A. The immunology of primary HIV infection: which immune responses control HIV replication? Curr. Opin. Immunol. 7, 456–461 ( 1995).
Koup, R.A. et al. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68, 4650– 4655 (1994).
Borrow, P., Lewicki, H., Hahn, B.H., Shaw, G.M. & Oldstone, M.B. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68, 6103– 6110 (1994).
Musey, L. et al. Cytotoxic T cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N. Engl. J. Med. 337, 1267–1274 ( 1997).
Cao, Y., Qin, L., Zhang, L., Safrit, J. & Ho, D.D. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332, 201–208 (1995).
Harrer, T. et al. Cytotoxic T lymphocytes in asymptomatic long-term nonprogressing HIV-1 infection. Breadth and specificity of the response and relation to in vivo viral quasispecies in a person with prolonged infection and low viral load. J. Immunol. 156, 2616– 2623 (1996).
Klein, M.R. et al. Kinetics of gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J. Exp. Med. 181, 1365–1372 (1995).
Carmichael, A., Jin, X., Sissons, P. & Borysiewicz, L. Quantitative analysis of the human immunodeficiency virus type 1 (HIV-1) specific cytotoxic T lymphocytes (CTL) response at different stages of HIV-1 infection: differential CTL responses to HIV-1 and Epstein-Barr virus in late disease. J.f Exp. Med. 177, 249–256 (1993).
Goulder, P.J. et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nature Med. 3, 212–217 (1997).
Borrow, P. et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nature Med. 3, 205– 211 (1997).
Phillips, R.E. et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354, 453–459 (1991).
Cheynier, R. et al. HIV and T cell expansion in splenic white pulps is accompanied by infiltration of HIV-specific cytotoxic T lymphoctyes. Cell 78, 373–387 (1994).
Ogg, G.S. et al. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279, 2103– 2105 (1998).
Pantaleo, G. et al. Accumulation of human immunodeficiency virus-specific cytotoxic T lymphocytes away from the predominant site of virus replication during primary infection. Eur. J. Immunol. 27, 3166– 3173 (1997).
Collins, K.L., Chen, B.K., Kalams, S.A., Walker, B.D. & Baltimore, D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphoctyes. Nature 391, 397–401 (1998).
Zinkernagel, R.M. & Hengartner, H. T-cell-mediated immunopathology versus direct cytolysis by virus: implications for HIV and AIDS. Immunol. Today 15, 262– 268 (1994).
Plata, F. et al. AIDS virus-specific cytotoxic T lymphocytes in lung disorders. Nature 328, 348–351 (1987).
Jassoy, C., Johnson, R.P., Navia, B.A., Worth, J. & Walker, B.D. Detection of a vigorous HIV-1-specific cytotoxic T lymphocyte response in the cerebrospinal fluid from infected persons with AIDS dementia complex. J. Immunol. 149, 3113–3119 (1992).
Heslop, H.E. et al. Long-term restoration of immunity against Epstein-Barr virus infection by adoptive transfer of gene-modified virus-specific T lymphocytes. Nature Med. 2, 551–555 (1996).
Riddell, S.R. et al. Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science 257 , 238–241 (1992).
Riddell, S.R., Gilbert, M.J. & Greenberg, P.D. CD8+ cytotoxic T cell therapy of cytomegalovirus and HIV infection. Curr. Opin. Immunol. 5, 484 –491 (1993).
Riddell, S.R. et al. T-cell mediated rejection of gene-modified HIV-specific cytotoxic T lymphocytes in HIV-infected patients. Nature Med. 2, 216–223 (1996).
Embretson, J. et al. Massive covert infection of helper T lymphocytes and macrophages by HIV during the incubation period of AIDS. Nature 362, 359–362 (1993).
Walker, R.E. et al. Peripheral expansion of pre-existing mature T cells is an important means of CD4+ T-cell regeneration HIV-infected adults. Nature Med. 4, 852–856 (1998).
Reddehase, M.J., Mutter, W. & Koszinowski, U.H. In vivo application of recombinant interleukin 2 in the immunotherapy of established cytomegalovirus infection. J. Exp. Med. 165, 650–656 (1987).
Matloubian, M., Concepcion, R.J. & Ahmed, R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J. Virol. 68, 8056–8063 (1994).
Rosenberg, E.S., et al. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 278, 1447– 1450 (1997).
Woffendin, C., Ranga, U., Yang, Z., Xu, L. & Nabel, G.J. Expression of a protective gene prolongs survival of T cells in human immunodeficiency virus-infected patients. Proc. Natl. Acad. Sci. USA 93, 2889–2894 (1996).
Nelson, B.H., Lord, J.D. & Greenberg, P.D. Cytoplasmic domains of the interleukin-2 receptor beta and gamma chains mediate the signal for T-cell proliferation. Nature 369, 333–336 ( 1994).
Patterson, B.K. et al. Detection of HIV-1 DNA and messenger RNA in individual cells by PCR-driven in situ hybridization and flow cytometry. Science 260, 976–979 ( 1993).
Patterson, B.K., Goolsby, C., Hodara, V., Lohman, K.L. & Wolinsky, S.M. Detection of CD4+ T cells harboring human immunodeficiency virus type 1 DNA by flow cytometry using simultaneous immunophenotyping and PCR-driven in situ hybridization: evidence of epitope masking of the CD4 cells surface molecule in vivo. J. Virol. 69, 4316–4322 (1995).
Patterson, B.K., Mosiman, V.L., Cantarero, L., Bhattacharya, M. & Goolsby, C. Detection of HIV-RNA-positive monocytes in peripheral blood of HIV-positive patients by simultaneous flow cytometric analysis of intracellular HIV RNA and cellular immunophenotype. Cytometry 31, 265–274 ( 1998).
Yang, L.P. et al. Productive infection of neonatal CD8+ T lymphocytes by HIV-1. J. Exp. Med. 187, 1139– 1144 (1998).
Brodie, S.J. et al. The effects of pharmacological and lentivirus-induced immune suppression on orbivirus pathogenesis: assessment of virus burden in blood monocytes and tissues by reverse transcription-in situ PCR. J. Virol. 72, 5599–5609 (1998).
Brodie, S.J. et al. Macrophage function in simian AIDS. Killing defects in vivo are independent of macrophage infection, associated with alterations in Th phenotype, and reversible with IFN-gamma. J. Immunol. 153, 5790–5801 (1994).
Brodie, S.J. et al. Epizootic hemorrhagic disease: analysis of tissues by amplification and in situ hybridization reveals widespread orbivirus infection at low copy numbers. J. Virol. 72, 3863– 3871 (1998).
Diamond, C. et al. Human herpesvirus 8 in the prostate glands of men with Kaposi's sarcoma. J. Virol. 72, 6223– 6227 (1998).
Brodie, S.J. et al. Ovine lentivirus expression and disease. Virus replication, but not entry, is restricted to macrophages of specific tissues. Am. J. Pathol. 146, 250–263 (1995).
Fox, C.H. & Cottler-Fox, M. In situ hybridization for detection of HIV RNA. Curr. Protocol. Immunol. 12.18.11–21 (1994).
Acknowledgements
We thank D. Spach (AIDS Clinical Trial Unit, University of Washington) for patient referral; K. Diem, M. Elliot, S. Friborg (Targeted Genetics), C. Hoffer, D. Kelly, C. B. Joiner, J. Joyce, V. Mosiman, D. Muthui, P.M. O'Hearn, E.L. Peterson, L. Wilson (Targeted Genetics) and B. Wood for technical assistance; B. Gilman and E. Gilman for their gift to the Program in Immunology; and Targeted Genetics for obtaining the IND for these studies, for providing the LN retroviral vector and for doing quality control testing. This study was supported by funds from the National Institutes of Health (AI36613 and AI41535 to S.B., P30 HD28834 and AI-27757 to D.L., AI30731 to S.R., P.D.G, and L.C.) and from the Pediatric AIDS Foundation, 77296-19—PF to D.L.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Brodie, S., Lewinsohn, D., Patterson, B. et al. In vivo migration and function of transferred HIV-1-specific cytotoxic T cells. Nat Med 5, 34–41 (1999). https://doi.org/10.1038/4716
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/4716