Abstract
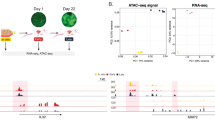
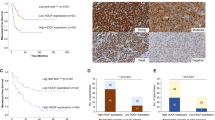
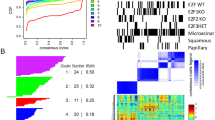
Metastasis results from a complex set of traits acquired by tumor cells, distinct from those necessary for tumorigenesis. Here, we investigate the contribution of enhancer elements to the metastatic phenotype of osteosarcoma. Through epigenomic profiling, we identify substantial differences in enhancer activity between primary and metastatic human tumors and between near isogenic pairs of highly lung metastatic and nonmetastatic osteosarcoma cell lines. We term these regions metastatic variant enhancer loci (Met-VELs). Met-VELs drive coordinated waves of gene expression during metastatic colonization of the lung. Met-VELs cluster nonrandomly in the genome, indicating that activity of these enhancers and expression of their associated gene targets are positively selected. As evidence of this causal association, osteosarcoma lung metastasis is inhibited by global interruptions of Met-VEL-associated gene expression via pharmacologic BET inhibition, by knockdown of AP-1 transcription factors that occupy Met-VELs, and by knockdown or functional inhibition of individual genes activated by Met-VELs, such as that encoding coagulation factor III/tissue factor (F3). We further show that genetic deletion of a single Met-VEL at the F3 locus blocks metastatic cell outgrowth in the lung. These findings indicate that Met-VELs and the genes they regulate play a functional role in metastasis and may be suitable targets for antimetastatic therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Change history
07 February 2018
In the version of this article initially published, two of the authors are incorrectly identified as John Stamatoyannopolus and Henri Versteeg. The authors' names are John A Stamatoyannopoulos and Henri H Versteeg. Also, the affiliation "Research Laboratory, Istituto Ortopedico Rizzoli, Bologna, Italy" is incorrect. The correct affiliation is "Laboratory of Experimental Oncology, Istituto Ortopedico Rizzoli, Bologna, Italy". The errors have been corrected in the HTML and PDF versions of the article.
References
Valastyan, S. & Weinberg, R.A. Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292 (2011).
Chambers, A.F., Groom, A.C. & MacDonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2, 563–572 (2002).
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015).
Hong, M.K. et al. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat. Commun. 6, 6605 (2015).
Bos, P.D. et al. Genes that mediate breast cancer metastasis to the brain. Nature 459, 1005–1009 (2009).
Kang, Y. et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 (2003).
Minn, A.J. et al. Genes that mediate breast cancer metastasis to lung. Nature 436, 518–524 (2005).
Factor, D.C. et al. Epigenomic comparison reveals activation of “seed” enhancers during transition from naive to primed pluripotency. Cell Stem Cell 14, 854–863 (2014).
Gifford, C.A. et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153, 1149–1163 (2013).
Zhu, J. et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell 152, 642–654 (2013).
Heintzman, N.D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009).
Akhtar-Zaidi, B. et al. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336, 736–739 (2012).
Cohen, A.J. et al. Hotspots of aberrant enhancer activity punctuate the colorectal cancer epigenome. Nat. Commun. 8, 14400 (2017).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Lovén, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Ramaswamy, S., Ross, K.N., Lander, E.S. & Golub, T.R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33, 49–54 (2003).
McDonald, O.G. et al. Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat. Genet. 49, 367–376 (2017).
Kansara, M., Teng, M.W., Smyth, M.J. & Thomas, D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer 14, 722–735 (2014).
Huang, Y.M., Hou, C.H., Hou, S.M. & Yang, R.S. The metastasectomy and timing of pulmonary metastases on the outcome of osteosarcoma patients. Clin. Med. Oncol. 3, 99–105 (2009).
Whyte, W.A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Ren, L. et al. Characterization of the metastatic phenotype of a panel of established osteosarcoma cells. Oncotarget 6, 29469–29481 (2015).
Zentner, G.E., Tesar, P.J. & Scacheri, P.C. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 21, 1273–1283 (2011).
Rada-Iglesias, A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011).
Huang, H., Bhat, A., Woodnutt, G. & Lappe, R. Targeting the ANGPT–TIE2 pathway in malignancy. Nat. Rev. Cancer 10, 575–585 (2010).
Clayton, P.E., Banerjee, I., Murray, P.G. & Renehan, A.G. Growth hormone, the insulin-like growth factor axis, insulin and cancer risk. Nat. Rev. Endocrinol. 7, 11–24 (2011).
Pinski, J. et al. Inhibition of growth of human osteosarcomas by antagonists of growth hormone-releasing hormone. J. Natl. Cancer Inst. 87, 1787–1794 (1995).
Li, N. et al. Phosphodiesterase 10A: a novel target for selective inhibition of colon tumor cell growth and β-catenin-dependent TCF transcriptional activity. Oncogene 34, 1499–1509 (2015).
van den Berg, Y.W., Osanto, S., Reitsma, P.H. & Versteeg, H.H. The relationship between tissue factor and cancer progression: insights from bench and bedside. Blood 119, 924–932 (2012).
Mendoza, A. et al. Modeling metastasis biology and therapy in real time in the mouse lung. J. Clin. Invest. 120, 2979–2988 (2010).
Corradin, O. et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 24, 1–13 (2014).
Leaner, V.D. et al. Inhibition of AP-1 transcriptional activity blocks the migration, invasion, and experimental metastasis of murine osteosarcoma. Am. J. Pathol. 174, 265–275 (2009).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Lamoureux, F. et al. Selective inhibition of BET bromodomain epigenetic signalling interferes with the bone-associated tumour vicious cycle. Nat. Commun. 5, 3511 (2014).
Puissant, A. et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 3, 308–323 (2013).
Bandopadhayay, P. et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin. Cancer Res. 20, 912–925 (2014).
Fellmann, C. et al. An optimized microRNA backbone for effective single-copy RNAi. Cell Rep. 5, 1704–1713 (2013).
Versteeg, H.H. et al. Inhibition of tissue factor signaling suppresses tumor growth. Blood 111, 190–199 (2008).
You, J.S. & Jones, P.A. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22, 9–20 (2012).
Jones, S. et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl. Acad. Sci. USA 105, 4283–4288 (2008).
Liu, W. et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat. Med. 15, 559–565 (2009).
Campbell, P.J. et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–1113 (2010).
Yachida, S. et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117 (2010).
Navin, N. et al. Tumour evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Moelans, C.B. et al. Genomic evolution from primary breast carcinoma to distant metastasis: Few copy number changes of breast cancer related genes. Cancer Lett. 344, 138–146 (2014).
Kerbel, R.S., Frost, P., Liteplo, R., Carlow, D.A. & Elliott, B.E. Possible epigenetic mechanisms of tumor progression: induction of high-frequency heritable but phenotypically unstable changes in the tumorigenic and metastatic properties of tumor cell populations by 5-azacytidine treatment. J. Cell. Physiol. Suppl. 3, 87–97 (1984).
Rodenhiser, D.I. Epigenetic contributions to cancer metastasis. Clin. Exp. Metastasis 26, 5–18 (2009).
Javaid, S. et al. Dynamic chromatin modification sustains epithelial-mesenchymal transition following inducible expression of Snail-1. Cell Rep. 5, 1679–1689 (2013).
Latil, M. et al. Cell-type-specific chromatin states differentially prime squamous cell carcinoma tumor-initiating cells for epithelial to mesenchymal transition. Cell Stem Cell 20, 191–204.e5 (2017).
Denny, S.K. et al. Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell 166, 328–342 (2016).
Roe, J.S. et al. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell 170, 875–888.e20 (2017).
Khanna, C. et al. An orthotopic model of murine osteosarcoma with clonally related variants differing in pulmonary metastatic potential. Clin. Exp. Metastasis 18, 261–271 (2000).
Schmidt, D. et al. ChIP–seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48, 240–248 (2009).
McLean, C.Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Reimand, J., Arak, T. & Vilo, J. g:Profiler—a web server for functional interpretation of gene lists (2011 update). Nucleic Acids Res. 39, W307–15 (2011).
Merico, D., Isserlin, R., Stueker, O., Emili, A. & Bader, G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5, e13984 (2010).
Song, L. & Crawford, G.E. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb. Protoc. https://doi.org/10.1101/pdb.prot5384 (2010).
van de Werken, H.J. et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat. Methods 9, 969–972 (2012).
Liu, T. et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12, R83 (2011).
Knott, S.R.V. et al. A computational algorithm to predict shRNA potency. Mol. Cell 56, 796–807 (2014).
Zuber, J. et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol. 29, 79–83 (2011).
Goecks, J., Nekrutenko, A. & Taylor, J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 (2010).
Osborne, T.S. et al. Evaluation of eIF4E expression in an osteosarcoma-specific tissue microarray. J. Pediatr. Hematol. Oncol. 33, 524–528 (2011).
Sakuma, T. et al. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 18, 315–326 (2013).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Acknowledgements
The authors thank the members of the Tesar, Kaplan, and Helman laboratories for their input throughout the course of the project as well as B. Decker for his input on the manuscript text. Additional support was provided by the Genomics Core Facility of the Case Western Reserve University School of Medicine's Genetics and Genome Sciences Department and the Case Comprehensive Cancer Center (P30CA043703). This work was supported by the Liddy Shriver Sarcoma Initiative (P.C.S., C.K., J.J.M.), the QuadW Foundation (P.C.S.), Sarcoma Foundation of America (P.C.S.), St. Baldrick's Foundation (A.Y.H.), Alex's Lemonade Stand Foundation (A.Y.H.), Hyundai Hope-on-Wheels Program (A.Y.H.), Pediatric Cancer Research Foundation (A.Y.H.), CCCC AYA Oncology Pilot Grant (A.Y.H.), National Institutes of Health (NIH) grants F30 CA186633 (J.J.M.), F30 CA183510 (T.E.M.), T32 GM007250 (J.J.M., T.E.M., S.H.), R01CA193677 (P.C.S.), R01CA204279 (P.C.S.), R01CA160356 (P.C.S.), F31CA192874 (F.A.), R21CA218790 (A.Y.H.), NIH Intramural Visiting Fellow Program 15335 (M.M.L.), and NIH Intramural Research Program (C.K.).
Author information
Authors and Affiliations
Contributions
J.J.M., C.K., and P.C.S. conceived the overall experimental design. J.J.M., C.F.B., and G.D. generated ChIP–seq, RNA-seq, and DHS-seq data. J.J.M., A.S., S.H., and P.C.S. completed analyses of ChIP–seq, RNA-seq, and DHS-seq data. J.J.M. and T.E.M. designed and completed the shRNA screening experiment and subsequent analysis. T.E.M. completed functional enrichment analysis of RNA-seq data. J.J.M. and I.B. generated 4C-seq data. J.J.M. and A.S. analyzed 4C-seq data. J.J.M., A.M., and I.B. and M.M.L. completed the in vivo and ex vivo metastasis experiments. J.J.M., J.T.M., and F.A. designed and completed the orthotopic metastasis experiments. J.J.M., D.R.C., and A.P.W.F. designed and completed the TALEN deletion experiments. M.Y.K. completed the in vitro F3 experiments. M.G., A.R., and P.P. provided subject tumor samples and clinical data. B.P.R. assessed F3 staining in subject tissue microarrays. A.D., A.Y.H., P.S.M., L.J.H., H.H.V., J.A.S., C.K., and P.C.S. provided the technical expertise and facilities to complete the experiments. J.J.M. and P.C.S. analyzed all data and wrote the paper. All authors provided intellectual input, edited, and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures & Tables
Supplementary Figures 1–13 and Supplementary Table 1 (PDF 2043 kb)
Supplementary Table 2
Met-VEL gene overlaps across patient tumors and cell lines (XLSX 468 kb)
Supplementary Table 3
Hairpins used in high-throughput in vivo RNAi screen (XLSX 37 kb)
Rights and permissions
About this article
Cite this article
Morrow, J., Bayles, I., Funnell, A. et al. Positively selected enhancer elements endow osteosarcoma cells with metastatic competence. Nat Med 24, 176–185 (2018). https://doi.org/10.1038/nm.4475
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4475
This article is cited by
-
POLE2 promotes osteosarcoma progression by enhancing the stability of CD44
Cell Death Discovery (2024)
-
Host-derived growth factors drive ERK phosphorylation and MCL1 expression to promote osteosarcoma cell survival during metastatic lung colonization
Cellular Oncology (2024)
-
Oncogenic enhancers prime quiescent metastatic cells to escape NK immune surveillance by eliciting transcriptional memory
Nature Communications (2024)
-
Etiology of super-enhancer reprogramming and activation in cancer
Epigenetics & Chromatin (2023)
-
Temporal chromatin accessibility changes define transcriptional states essential for osteosarcoma metastasis
Nature Communications (2023)