Abstract
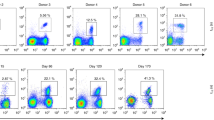
Cytotoxic T lymphocytes (CTLs) are critical for the control of human immunodeficiency virus, but containment of virus replication can be undermined by mutations in CTL epitopes that lead to virus escape. We analyzed the evolution in vivo of an immunodominant, HLA-A2–restricted CTL epitope and found two principal, diametrically opposed evolutionary pathways that exclusively affect T cell–receptor contact residues. One pathway was characterized by acquisition of CTL escape mutations and the other by selection for wild-type amino acids. The pattern of CTL responses to epitope variants shaped which variant(s) prevailed in the virus population. The pathways notably influenced the amount of plasma virus, as patients with efficient CTL selection had lower plasma viral loads than did patients without efficient selection. Thus, viral escape from CTL responses does not necessarily correlate with disease progression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
McMichael, A. & Klenerman, P. HIV/AIDS. HLA leaves its footprints on HIV. Science 296, 1410–1411 (2002).
Moore, C.B. et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296, 1439–1443 (2002).
Brander, C. & Walker, B.D. Gradual adaptation of HIV to human host populations: good or bad news? Nat Med. 9, 1359–1362 (2003).
Allen, T.M. et al. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407, 386–390 (2000).
Borrow, P. et al. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3, 205–211 (1997).
Price, D.A. et al. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94, 1890–1895 (1997).
Jamieson, B.D. et al. Epitope escape mutation and decay of human immunodeficiency virus type 1-specific CTL responses. J. Immunol. 171, 5372–5379 (2003).
Kelleher, A.D. et al. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193, 375–386 (2001).
Leslie, A.J. et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10, 282–289 (2004).
Altfeld, M. et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17, 2581–2591 (2003).
Goulder, P.J. et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3, 212–217 (1997).
Altfeld, M. et al. The majority of currently circulating human immunodeficiency virus type 1 clade B viruses fail to prime cytotoxic T-lymphocyte responses against an otherwise immunodominant HLA-A2-restricted epitope: implications for vaccine design. J. Virol. 79, 5000–5005 (2005).
Goulder, P.J. et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412, 334–338 (2001).
Browning, M. & Krausa, P. Genetic diversity of HLA-A2: evolutionary and functional significance. Immunol. Today 17, 165–170 (1996).
Johnson, R.P. et al. HIV-1 gag-specific cytotoxic T lymphocytes recognize multiple highly conserved epitopes. Fine specificity of the gag-specific response defined by using unstimulated peripheral blood mononuclear cells and cloned effector cells. J. Immunol. 147, 1512–1521 (1991).
Tsomides, T.J. et al. Naturally processed viral peptides recognized by cytotoxic T lymphocytes on cells chronically infected by human immunodeficiency virus type 1. J. Exp. Med. 180, 1283–1293 (1994).
HIV Sequence Compendium 2003 (eds. Leitner, T. et al.) LA-UR 04-7420 (Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, New Mexico, USA, 2004).
Ogg, G.S. et al. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279, 2103–2106 (1998).
Goulder, P.J. et al. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193, 181–194 (2001).
Ogg, G.S. et al. Longitudinal phenotypic analysis of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes: correlation with disease progression. J. Virol. 73, 9153–9160 (1999).
Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997).
Goulder, P.J. et al. Rapid definition of five novel HLA-A*3002-restricted human immunodeficiency virus-specific cytotoxic T-lymphocyte epitopes by elispot and intracellular cytokine staining assays. J. Virol. 75, 1339–1347 (2001).
Brander, C. et al. Lack of strong immune selection pressure by the immunodominant, HLA-A*0201-restricted cytotoxic T lymphocyte response in chronic human immunodeficiency virus-1 infection. J. Clin. Invest. 101, 2559–2566 (1998).
Sette, A. et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 153, 5586–5592 (1994).
Lee, J.K. et al. T cell cross-reactivity and conformational changes during TcR engagement. J. Exp. Med. 200, 1455–1466 (2004).
van der Merwe, P.A. & Davis, S.J. Molecular interactions mediating T cell antigen recognition. Annu. Rev. Immunol. 21, 659–684 (2003).
Madden, D.R., Garboczi, D.N. & Wiley, D.C. The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell 75, 693–708 (1993).
Ding, Y.H. et al. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity 8, 403–411 (1998).
Garboczi, D.N. et al. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature 384, 134–141 (1996).
Stewart-Jones, G.B., McMichael, A.J., Bell, J.I., Stuart, D.I. & Jones, E.Y. A structural basis for immunodominant human T cell receptor recognition. Nat. Immunol. 4, 657–663 (2003).
Edwards, C.T., Pfafferott, K.J., Goulder, P.J., Phillips, R.E. & Holmes, E.C. Intrapatient escape in the A*0201-restricted epitope SLYNTVATL drives evolution of human immunodeficiency virus type 1 at the population level. J. Virol. 79, 9363–9366 (2005).
Feeney, M.E. et al. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78, 8927–8930 (2004).
Goulder, P.J. et al. Combined structural and immunological refinement of HIV-1 HLA-B8-restricted cytotoxic T lymphocyte epitopes. Eur. J. Immunol. 27, 1515–1521 (1997).
Yokomaku, Y. et al. Impaired processing and presentation of cytotoxic-T-lymphocyte (CTL) epitopes are major escape mechanisms from CTL immune pressure in human immunodeficiency virus type 1 infection. J. Virol. 78, 1324–1332 (2004).
Matano, T. et al. Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. J. Exp. Med. 199, 1709–1718 (2004).
Friedrich, T.C. et al. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10, 275–281 (2004).
Kan-Mitchell, J. et al. The HIV-1 HLA-A2-SLYNTVATL is a help-independent CTL epitope. J. Immunol. 172, 5249–5261 (2004).
Sun, J.C. & Bevan, M.J. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science 300, 339–342 (2003).
Shedlock, D.J. & Shen, H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science 300, 337–339 (2003).
Krogsgaard, M. et al. Evidence that structural rearrangements and/or flexibility during TCR binding can contribute to T cell activation. Mol. Cell 12, 1367–1378 (2003).
Iversen, A.K. et al. Cervical human immunodeficiency virus type 1 shedding is associated with genital beta-chemokine secretion. J. Infect. Dis. 178, 1334–1342 (1998).
Iversen, A.K. et al. Distinct determinants of human immunodeficiency virus type 1 RNA and DNA loads in vaginal and cervical secretions. J. Infect. Dis. 177, 1214–1220 (1998).
Iversen, A.K. et al. Presence of multiple HIV subtypes and a high frequency of subtype chimeric viruses in heterosexually infected women. J. Acquir. Immune Defic. Syndr. 22, 325–332 (1999).
Brander, C. et al. Efficient processing of the immunodominant, HLA-A*0201-restricted human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitope despite multiple variations in the epitope flanking sequences. J. Virol. 73, 10191–10198 (1999).
Goulder, P.J. et al. Mother-to-child transmission of HIV infection and CTL escape through HLA-A2-SLYNTVATL epitope sequence variation. Immunol. Lett. 79, 109–116 (2001).
Goulder, P.J. et al. Patterns of immunodominance in HIV-1-specific cytotoxic T lymphocyte responses in two human histocompatibility leukocyte antigens (HLA)-identical siblings with HLA-A*0201 are influenced by epitope mutation. J. Exp. Med. 185, 1423–1433 (1997).
Sylvester-Hvid, C. et al. Establishment of a quantitative ELISA capable of determining peptide - MHC class I interaction. Tissue Antigens 59, 251–258 (2002).
Dong, T. et al. An HLA-B35-restricted epitope modified at an anchor residue results in an antagonist peptide. Eur. J. Immunol. 26, 335–339 (1996).
Garboczi, D.N., Madden, D.R. & Wiley, D.C. Five viral peptide-HLA-A2 co-crystals. Simultaneous space group determination and X-ray data collection. J. Mol. Biol. 239, 581–587 (1994).
Willcox, B.E. et al. TCR binding to peptide-MHC stabilizes a flexible recognition interface. Immunity 10, 357–365 (1999).
Parham, P. & Brodsky, F.M. Partial purification and some properties of BB7.2. A cytotoxic monoclonal antibody with specificity for HLA-A2 and a variant of HLA-A28. Hum. Immunol. 3, 277–299 (1981).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Maddison, W.P. & Maddison, D.R. MacClade - Analysis of Phylogeny and Character Evolution - Version 4, (Sinauer Associates, Inc., Sunderland, MA, 2001).
Learn, G.H., Jr. et al. Maintaining the integrity of human immunodeficiency virus sequence databases. J. Virol. 70, 5720–5730 (1996).
Rose, P.P. & Korber, B.T. Detecting hypermutations in viral sequences with an emphasis on G -> A hypermutation. Bioinformatics 16, 400–401 (2000).
Iversen, A.K. Genital HIV shedding in women. AIDS Patient Care STDS 13, 695–701 (1999).
Huelsenbeck, J.P. & Crandall, K.A. Phylogeny estimation and hypothesis testing using maximum likelihood. Annu. Rev. Ecol. Syst. 28, 437–466 (1997).
Posada, D. & Crandall, K.A. MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818 (1998).
Swofford, D.L. PAUP* 4.0: Phylogenetic Analysis Using Parsimony (*and Other Methods) 4.0b2a edn. (Sinauer Associates, Sunderland, Massachusetts, 1999).
Akaike, H. A new look at statistical model identification. IEEE Trans. Autom. Contr. 19, 716–723 (1974).
Yang, Z. Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza virus A. J. Mol. Evol. 51, 423–432 (2000).
Nielsen, R. & Yang, Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 148, 929–936 (1998).
Hill, C.P., Worthylake, D., Bancroft, D.P., Christensen, A.M. & Sundquist, W.I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl Acad. Sci. USA 93, 3099–3104 (1996).
Acknowledgements
We thank patients for donating samples, N. Willcox for comments on the manuscript, P. Goulder for discussions and C. Siebold for assistance with crystallographic refinement. Supported by the Novo Nordisk Foundation, the Danish AIDS foundation, the University of Washington Center for AIDS Research, International AIDS Vaccine Initiative, Cancer Research UK (E.Y.J.) and Medical Research Council UK (A.K.N.I. and A.J.M.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Additional clinical and sampling data (PDF 17 kb)
Supplementary Table 2
Crystallographic data (PDF 27 kb)
Rights and permissions
About this article
Cite this article
Iversen, A., Stewart-Jones, G., Learn, G. et al. Conflicting selective forces affect T cell receptor contacts in an immunodominant human immunodeficiency virus epitope. Nat Immunol 7, 179–189 (2006). https://doi.org/10.1038/ni1298
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni1298
This article is cited by
-
Impact of pre-adapted HIV transmission
Nature Medicine (2016)
-
HIV-infected sex workers with beneficial HLA-variants are potential hubs for selection of HIV-1 recombinants that may affect disease progression
Scientific Reports (2015)
-
Definition of the viral targets of protective HIV-1-specific T cell responses
Journal of Translational Medicine (2011)
-
Unique features of HLA-mediated HIV evolution in a Mexican cohort: a comparative study
Retrovirology (2009)
-
Antigen-processing and presentation pathways select antigenic HIV peptides in the fight against viral evolution
Nature Immunology (2009)