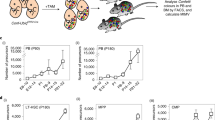
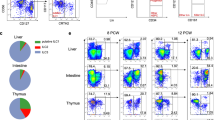
Abstract
The molecular events that initiate lymphoid-lineage specification remain unidentified because the stages of differentiation during which lineage commitment occurs are difficult to characterize. We isolated fetal liver progenitor cells undergoing restriction of their differentiation potential toward the T cell–innate lymphoid cell lineage or the B cell lineage. Transcripts that defined the molecular signatures of these two subsets were sequentially upregulated in lympho-myeloid precursor cells and in common lymphoid progenitor cells, respectively, and this preceded lineage restriction; this indicates that T cell–versus–B cell commitment is not a binary fate 'decision'. The T cell–bias and B cell–bias transcriptional programs were frequently co-expressed in common lymphoid progenitor cells and were segregated in subsets biased toward T cell differentiation or B cell differentiation, after interleukin 7 (IL-7) signaling that controlled the number of progenitor cells engaging in T cell differentiation versus B cell differentiation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Nutt, S.L. & Kee, B.L. The transcriptional regulation of B cell lineage commitment. Immunity 26, 715–725 (2007).
Rothenberg, E.V., Moore, J.E. & Yui, M.A. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 8, 9–21 (2008).
Kondo, M., Weissman, I.L. & Akashi, K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell 91, 661–672 (1997).
Adolfsson, J. et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell 121, 295–306 (2005).
Mebius, R.E. et al. The fetal liver counterpart of adult common lymphoid progenitors gives rise to all lymphoid lineages, CD45+CD4+CD3− cells, as well as macrophages. J. Immunol. 166, 6593–6601 (2001).
Karsunky, H., Inlay, M.A., Serwold, T., Bhattacharya, D. & Weissman, I.L. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood 111, 5562–5570 (2008).
Pereira de Sousa, A. et al. Inhibitors of DNA binding proteins restrict T cell potential by repressing Notch1 expression in Flt3-negative common lymphoid progenitors. J. Immunol. 189, 3822–3830 (2012).
Yoshida, H. et al. Expression of α4β7 integrin defines a distinct pathway of lymphoid progenitors committed to T cells, fetal intestinal lymphotoxin producer, NK, and dendritic cells. J. Immunol. 167, 2511–2521 (2001).
Possot, C. et al. Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat. Immunol. 12, 949–958 (2011).
Rieger, M.A., Hoppe, P.S., Smejkal, B.M., Eitelhuber, A.C. & Schroeder, T. Hematopoietic cytokines can instruct lineage choice. Science 325, 217–218 (2009).
Miller, J.P. et al. The earliest step in B lineage differentiation from common lymphoid progenitors is critically dependent upon interleukin 7. J. Exp. Med. 196, 705–711 (2002).
Peschon, J.J. et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 180, 1955–1960 (1994).
Sudo, T. et al. Interleukin 7 production and function in stromal cell-dependent B cell development. J. Exp. Med. 170, 333–338 (1989).
Schlissel, M.S., Durum, S.D. & Muegge, K. The interleukin 7 receptor is required for T cell receptor gamma locus accessibility to the V(D)J recombinase. J. Exp. Med. 191, 1045–1050 (2000).
Kikuchi, K., Lai, A.Y., Hsu, C.-L. & Kondo, M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J. Exp. Med. 201, 1197–1203 (2005).
Dias, S., Silva, H. Jr., Cumano, A. & Vieira, P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J. Exp. Med. 201, 971–979 (2005).
Tsapogas, P. et al. IL-7 mediates Ebf-1-dependent lineage restriction in early lymphoid progenitors. Blood 118, 1283–1290 (2011).
Malin, S. et al. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat. Immunol. 11, 171–179 (2010).
Carvalho, T.L., Mota-Santos, T., Cumano, A., Demengeot, J. & Vieira, P. Arrested B lymphopoiesis and persistence of activated B cells in adult interleukin 7−/− mice. J. Exp. Med. 194, 1141–1150 (2001).
Vosshenrich, C.A.J., Cumano, A., Müller, W., Di Santo, J.P. & Vieira, P. Thymic stromal-derived lymphopoietin distinguishes fetal from adult B cell development. Nat. Immunol. 4, 773–779 (2003).
Borge, O.J., Adolfsson, J., Mårtensson, A., Mårtensson, I.L. & Jacobsen, S.E. Lymphoid-restricted development from multipotent candidate murine stem cells: distinct and complimentary functions of the c-kit and flt3-ligands. Blood 94, 3781–3790 (1999).
Sitnicka, E. et al. Critical role of FLT3 ligand in IL-7 receptor independent T lymphopoiesis and regulation of lymphoid-primed multipotent progenitors. Blood 110, 2955–2964 (2007).
Bhandoola, A., Sambandam, A., Allman, D., Meraz, A. & Schwarz, B. Early T lineage progenitors: new insights, but old questions remain. J. Immunol. 171, 5653–5658 (2003).
Ramond, C. et al. Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nat. Immunol. 15, 27–35 (2014).
Porritt, H.E. et al. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity 20, 735–745 (2004).
Inlay, M.A. et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 23, 2376–2381 (2009).
Luc, S. et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat. Immunol. 13, 412–419 (2012).
Lin, H. & Grosschedl, R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376, 263–267 (1995).
Nutt, S.L., Heavey, B., Rolink, A.G. & Busslinger, M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401, 556–562 (1999).
Desanti, G.E. et al. Clonal analysis reveals uniformity in the molecular profile and lineage potential of CCR9(+) and CCR9(−) thymus-settling progenitors. J. Immunol. 186, 5227–5235 (2011).
Masuda, K. et al. Prethymic T-cell development defined by the expression of paired immunoglobulin-like receptors. EMBO J. 24, 4052–4060 (2005).
Lee, D.M. et al. The mouse CD7 gene: identification of a new element common to the human CD7 and mouse Thy-1 promoters. Immunogenetics 44, 108–114 (1996).
Senée, V. et al. Mutations in GLIS3 are responsible for a rare syndrome with neonatal diabetes mellitus and congenital hypothyroidism. Nat. Genet. 38, 682–687 (2006).
Del Real, M.M. & Rothenberg, E.V. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development 140, 1207–1219 (2013).
Weber, B.N. et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68 (2011).
Han, H. et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14, 637–645 (2002).
Schlenner, S.M. et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity 32, 426–436 (2010).
Uehara, S., Grinberg, A., Farber, J.M. & Love, P.E. A role for CCR9 in T lymphocyte development and migration. J. Immunol. 168, 2811–2819 (2002).
Lai, A.Y. & Kondo, M. Identification of a bone marrow precursor of the earliest thymocytes in adult mouse. Proc. Natl. Acad. Sci. USA 104, 6311–6316 (2007).
Igarashi, H., Gregory, S.C., Yokota, T., Sakaguchi, N. & Kincade, P.W. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity 17, 117–130 (2002).
Changelian, P.S. et al. Prevention of organ allograft rejection by a specific Janus kinase 3 inhibitor. Science 302, 875–878 (2003).
Cvejic, A. Mechanisms of fate decision and lineage commitment during haematopoiesis. Immunol. Cell Biol. 94, 230–235 (2016).
Chea, S. et al. Single-cell gene expression analyses reveal heterogeneous responsiveness of fetal innate lymphoid progenitors to Notch signaling. Cell Rep. 14, 1500–1516 (2016).
McKenna, H.J. et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood 95, 3489–3497 (2000).
von Freeden-Jeffry, U. et al. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181, 1519–1526 (1995).
Reich, M. et al. GenePattern 2.0. Nat. Genet. 38, 500–501 (2006).
Eisen, M.B., Spellman, P.T., Brown, P.O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868 (1998).
de Hoon, M.J.L., Imoto, S., Nolan, J. & Miyano, S. Open source clustering software. Bioinformatics 20, 1453–1454 (2004).
Heng, T.S.P., Painter, M.W. Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Acknowledgements
We thank A. Bandeira and D. Guy-Grand for critical reading; S. Novault and S. Schmutz for support; H.-R. Rodewald (German Cancer Center DKFZ) for the IL-7Rα-Cre mouse line; and P. Chambon (IGBMC Strasbourg) for the TSLP-KO mice. This work benefited from data assembled by the ImmGen Consortium. Supported by the Pasteur Institute, Institut National de la Santé et de la Recherche Médicale (INSERM), the Ministère de la Recherche (C.B., C.R. and S.C.), Association pour la Recherche sur le Cancer (S.C. and R.G.), La Ligue Contre Le Cancer (C.B. and C.R.), Université Paris Diderot (C.B., R.G. and S.C.), Université Pierre et Marie Curie (C.R.), the Agence Nationale de la Recherche (project Myeloten (R.G.); program REVIVE (Investment for the Future) (A.C.); and project Twothyme (A.C.) and the Pasteur-Weizmann Foundation (A.C.).
Author information
Authors and Affiliations
Contributions
C.B. designed and performed most experiments, analyzed data and wrote the manuscript; C.R., O.B.-D. and S.C. performed experiments; G.S. performed the microarray; R.G. provided mouse lines and designed experiments; C.R., P.P. and P.V. contributed to the discussions and writing; and A.C. directed the research, designed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Kinetics for the expression of HSA and Ly6D in fetal and adult lymphoid progenitor cells.
(a) E12, E13, E15 and E17 FL cells were stained as described in Fig 1 and HSA expression was analyzed in the iCLP (Flt3+ α4β7-) compartment. (b) Absolute numbers of HSAlo, HSAint and HSAhi iCLPs per E13, E15 and E16 fetal livers. Data is representative of at least 3 independent experiments. (c) Plots of iCLP from E13, E15 and E17 FL showing the HSA and Ly6D expressions (high and intermediate levels of HSA are marked). (d) Plots show the expression of Ly6D in BM Flt3+ IL7Rα+ CLP. (e) Venn diagrams representing the number of overlapping upregulated (left) and downregulated (right) transcripts in the HSAhi, HSAlo and α4β7 CLP compared to HSAintiCLPs. (f) Quantitative RT-PCR gene expression analysis of the HSAint-signature gene Eltd1 in HSAlo, HSAint, HSAhi iCLPs and HSC (Lin- c-Kit+ Sca-1+ Flt3-). Histograms display relative quantity of expression normalized to Hprt. Data is representative of 3 independent experiments. (g) Histogram displays the frequency of Eltd1 expressing single cells in HSAint iCLPs expressing neither ETP nor proB specific genes (None), expressing Pax5 or Pira11, and HSAhi and HSAlo iCLPs. Data represents a pool of 5 independent experiments of 14 single cells each. (f, g) mean±SEM **p<0.01***p<0.001****p<0.0001 (Student’s t test).
Supplementary Figure 2 Deletion of RBPJ in IL-7rα-expressing cells does not affect the B cell potential or expression of the T cell–biased transcriptional signature of HSAlo iCLPs.
(a) E14 CLPs isolated from (IL7RαCre/+ ROSAYFP) RBPJfx/+ (top panel) and RBPJfx/fx(bottom panel) littermates embryos were stained as described in Fig 1. PIRA/B expression was analyzed in α4β7- HSAlo and HSAintCLPs, stained as described in Fig 1. Data is representative of more than 5 independent experiments. (b) Absolute numbers of HSAlo, HSAint and HSAhi iCLP per E14 FL of RBPJfx/+ and RBPJfx/fx. Dot plots displays absolute CLP numbers in 5 embryos from 3 independent experiments, each dot represents a single embryo. (c) B cell differentiation potential of single HSAlo, HSAint and HSAhi iCLPs from E14 RBPJfx/+ and RBPJfx/fx was assessed as described in Fig 1. Data is representative of a pool of 3 independent experiments. (d) qRT-PCR gene expression analysis of HSAhi signature- genes Ebf1, Pax5, Hes1 (top panel) and of HSAlo signature-genes Glis3, Pira11, Cd7 (bottom panel) in HSAlo, HSAint and HSAhi iCLPs from E14 RBPJfx/+ and RBPJfx/fx. Histograms display relative quantity of expression normalized to Hprt. Data is a pool of 3 independent experiments. (e) Plots showing CD4 and CD19 expression in the progeny of α4β7- CLP IL7cre:RBPJfx/+ and RBPJfx/fx in culture for 10 days with OP9dll4, IL7, KitL and Flt3L. (b, c, d) mean±SEM *p<0.05 N.S. non significant (Student’s t test).
Supplementary Figure 3 Single-cell multiplex qPCR analysis of T cell–, B cell– and ILC-biased transcripts in FL CLPs.
Hierarchical unsupervised clustering of the multiplex single cell transcriptional analysis of 17 single HSAlo, HSAint and HSAhi iCLP and α4β7+ CLP for 28 transcripts of the HSAlo, HSAhi and α4β7+ CLP signatures and of Eltd1.
Supplementary Figure 4 PIRA/B-expressing FL CLPs and CRLPs co-express all T cell–bias genes similar to DN1a fetal thymocytes.
(a) E13 fetal blood and (b) thymocytes were isolated and stained for Lin (GR1, TER119, CD3ε, CD4, CD8, CD19, CD11c and NK1.1) CD44 CD25, c-kit, HSA and PIRA/B. DN1 were defined as Lin- CD44+ CD25- c-Kit+ and divided into DN1a (HSA-) and DN1b (HSA+), DN2 were defined as Lin- CD44+ CD25+ c-Kit+, and PIRA/B expression was analyzed in DN1a, DN1b and DN2. Single cell multiplex qRT-PCR on single sorted E13 (c) α4β7- HSAint PIRA/B+ CLPs (far left pannel), α4β7- HSAlo PIRA/B+ CLPs (middle left panel), α4β7- HSAlo PIRA/B+ CRLPs (middle right panel) and DN1a PIRA/B+ thymocytes (far right panel). The same set of 30 genes as in Fig 6 was analyzed by qRT-PCR using the Biomark Fluidigm system after 20 cycles of specific pre-amplification. Expression of Hprt, Pira11, Cd7, Ccr9, Glis3 and Ebf1 is displayed. Each dot represent detectable expression of a given gene and each horizontal line represent gene expression pattern in a given single cell. Frequency and absolute number of single cells expressing a given gene are displayed at the top of each column. (d) Plot showing the Ct values of Gata3 expression in HSAhi, HSAint and HSAlo PIR+ and PIR- CLP and in CRLP. Data is representative of 2 independent experiments. (d) mean±SEM **p<0.01 N.S. non significant (Student’s t test).
Supplementary Figure 5 PIRA/B+ T cell–biased LMPPs differentiate into PIRA/B+ T cell–biased iCLPs.
(a) Histogram displays frequency of single Flt3hi LMPP expressing Il7ra transcripts in WT and Il7-/- FL13. Data is representative of 82 to 89 single cells analyzed in 2 independent experiments. (b) Ct values of Il7ra transcripts in WT LMPP that express or not Ebf1 compared to iCLP. (c) Post sorting purity controls of FL E13 Il7-/- PIRA/B- (top panel) and PIRA/B+ (bottom panel) Flt3hi LMPPs. (d) Dot plots display the phenotype of the IL7Rα- compartment of cultured FL13 Il7-/- PIRA/B- and PIRA/B+ Flt3hi LMPPs after 24 hours of culture with saturating amounts of Flt3L and c-KitL, in the absence of stromal cells. Data is representative of 3 independent experiments. (a,b) mean±SEM. ****p<0.0001 (Student’s t test).
Supplementary Figure 6 IL-7 deficiency does not affect the proliferation, survival or B cell potential of E13 FL CLPs.
(a) Absolute numbers of CLPs per E13 FL in WT and Il7-/-embryos. Data is representative of 9 embryos analyzed in 3 independent experiments, each dot represents a single embryo. (b) qRT-PCR gene expression analysis of the HSAhi signature-genes Ebf1, Pax5, Hes1, Foxo1 and Rag2, of Notch1, and HSAlo signature-genes Glis3 and Pira11 in HSAlo, HSAint and HSAhi iCLPs from WT and Il7-/-embryos. Histograms display relative quantity of expression normalized to Hprt. Data is representative of 3 independent experiments. (c) Histogram displays frequency of B-lineage positive wells after culture of HSAint and HSAhi iCLPs from WT and Il7-/-embryos. Data is representative of a pool of 3 independent experiments. (d) Single cell multiplex qRT-PCR on single sorted E13 WT and Il7-/-HSAhi iCLPs, expression of Hprt, Ebf1, Pax5, Pira11, Cd7 and Glis3 is displayed. Data is representative of 3 independent experiments. (e,f) KI67 and DAPI staining and frequency of sorted HSAint and HSAlo iCLPs in S/G2/M (KI67+ DAPI+). Data is representative of 2 independent experiments. (g) Histogram display Annexin V staining on sorted HSAlo, HSAint and HSAhi WT (red line) and Il7-/-(blue line) iCLPs compared to adult thymocytes (gray tinted line). Data is representative of 2 independent experiments. (h) qRT-PCR gene expression analysis of Bcl2 transcripts in HSAlo, HSAint and HSAhi WT and Il7-/-iCLPs. Histograms display relative quantity of expression normalized to Hprt. Data is representative of 4 independent experiments. (a,b,c,f,h) mean±SEM *p<0.01 N.S. non significant (Student’s t test).
Supplementary Figure 7 Flt3 deficiency does not affect the commitment or priming of FL CLPs.
(a) WT and Flt3l-/- E13 FL were stained as described in Fig 1 and Flt3+ α4β7- CLP compartments were analyzed. Data is representative of 4 independent experiments. (b) Dot plots display PIRA/B expression in WT (left panel) and Flt3l-/-right panel) LSK compartment. Data is representative of 3 independent experiments. (c) Histograms display absolute numbers of total CLPs and HSAlo, HSAint and HSAhi iCLP fractions per E13 FL in WT and Flt3l-/-embryos. (d) Single cell multiplex qRT-PCR on single sorted E13 Flt3l-/- HSAint iCLPs, experiment was performed as described in Fig 3. Expression of Hprt, Ebf1, Pax5, Pira11, Cd7 and Glis3 is displayed. Data is representative of a pool of 2 independent experiments. (e) Display frequencies of single cell expressing T only, B only or T and B genes in Flt3l-/- HSAint iCLPs. “None” includes no B and no T gene expression. (f) Il7-/- and Il7-/-/Tslp-/- double KO E13 FL were stained as described in Fig 1 and Flt3+ α4β7- CLP compartments were analyzed. (g) Histograms display absolute numbers of HSAlo, HSAint and HSAhi iCLP fractions per E13 FL in Il7-/- and Il7-/-/Tslp-/-double KO embryos. Data is representative of 4 embryos per group analyzed independently. (c,g) mean±SEM **p<0.01***p<0.001 N.S. non significant (Student’s t test).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1–6.
Rights and permissions
About this article
Cite this article
Berthault, C., Ramond, C., Burlen-Defranoux, O. et al. Asynchronous lineage priming determines commitment to T cell and B cell lineages in fetal liver. Nat Immunol 18, 1139–1149 (2017). https://doi.org/10.1038/ni.3820
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3820