Abstract
Healthy individuals of African ancestry have neutropenia that has been linked with the variant rs2814778(G) of the gene encoding atypical chemokine receptor 1 (ACKR1). This polymorphism selectively abolishes the expression of ACKR1 in erythroid cells, causing a Duffy-negative phenotype. Here we describe an unexpected fundamental role for ACKR1 in hematopoiesis and provide the mechanism that links its absence with neutropenia. Nucleated erythroid cells had high expression of ACKR1, which facilitated their direct contact with hematopoietic stem cells. The absence of erythroid ACKR1 altered mouse hematopoiesis including stem and progenitor cells, which ultimately gave rise to phenotypically distinct neutrophils that readily left the circulation, causing neutropenia. Individuals with a Duffy-negative phenotype developed a distinct profile of neutrophil effector molecules that closely reflected the one observed in the ACKR1-deficient mice. Thus, alternative physiological patterns of hematopoiesis and bone marrow cell outputs depend on the expression of ACKR1 in the erythroid lineage, findings with major implications for the selection advantages that have resulted in the paramount fixation of the ACKR1 rs2814778(G) polymorphism in Africa.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Rot, A. & von Andrian, U.H. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu. Rev. Immunol. 22, 891–928 (2004).
Griffith, J.W., Sokol, C.L. & Luster, A.D. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu. Rev. Immunol. 32, 659–702 (2014).
Luther, S.A. & Cyster, J.G. Chemokines as regulators of T cell differentiation. Nat. Immunol. 2, 102–107 (2001).
Krathwohl, M.D. & Kaiser, J.L. Chemokines promote quiescence and survival of human neural progenitor cells. Stem Cells 22, 109–118 (2004).
Ulvmar, M.H., Hub, E. & Rot, A. Atypical chemokine receptors. Exp. Cell Res. 317, 556–568 (2011).
Bachelerie, F. et al. New nomenclature for atypical chemokine receptors. Nat. Immunol. 15, 207–208 (2014).
Nibbs, R.J.B. & Graham, G.J. Immune regulation by atypical chemokine receptors. Nat. Rev. Immunol. 13, 815–829 (2013).
Bachelerie, F. et al. Update on the extended family of chemokine receptors and introducing a new nomenclature for atypical chemokine receptors. Pharmacol. Rev. 66, 1–79 (2013).
Pruenster, M. et al. The Duffy antigen receptor for chemokines transports chemokines and supports their pro-migratory activity. Nat. Immunol. 10, 101–108 (2009).
Ulvmar, M.H. et al. The atypical chemokine receptor CCRL1 shapes functional CCL21 gradients in lymph nodes. Nat. Immunol. 15, 623–630 (2014).
Lee, K.M. et al. The chemokine receptors ACKR2 and CCR2 reciprocally regulate lymphatic vessel density. EMBO J. 33, 2564–2580 (2014).
Cruz-Orengo, L. et al. CXCR7 influences leukocyte entry into the CNS parenchyma by controlling abluminal CXCL12 abundance during autoimmunity. J. Exp. Med. 208, 327–339 (2011).
Novitzky-Basso, I. & Rot, A. Duffy antigen receptor for chemokines and its involvement in patterning and control of inflammatory chemokines. Front. Immunol. 3, 266 (2012).
Rot, A. Contribution of Duffy antigen to chemokine function. Cytokine Growth Factor Rev. 16, 687–694 (2005).
Mei, J. et al. CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity 33, 106–117 (2010).
Schnabel, R.B. et al. Duffy antigen receptor for chemokines (Darc) polymorphism regulates circulating concentrations of monocyte chemoattractant protein 1 and other inflammatory mediators. Blood 115, 5289–5299 (2010).
Reutershan, J., Harry, B., Chang, D., Bagby, G.J. & Ley, K. DARC on RBC limits lung injury by balancing compartmental distribution of CXC chemokines. Eur. J. Immunol. 39, 1597–1607 (2009).
Miller, L.H., Mason, S.J., Dvorak, J.A., McGinniss, M.H. & Rothman, I.K. Erythrocyte receptors for (Plasmodium knowlesi) malaria: Duffy blood group determinants. Science 189, 561–563 (1975).
Horuk, R. et al. A receptor for the malarial parasite Plasmodium vivax: the erythrocyte chemokine receptor. Science 261, 1182–1184 (1993).
Nedelec, Y. et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell 167, 657–669 (2016).
Quach, H. et al. Genetic adaptation and neandertal admixture shaped the immune system of human populations. Cell 167, 643–656 (2016).
Thobakgale, C.F. & Ndung'u, T. Neutrophil counts in persons of African origin. Curr. Opin. Hematol. 21, 50–57 (2014).
Reich, D. et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 5, e1000360 (2009).
Sanger, R., Race, R.R. & Jack, J. The Duffy blood groups of New York negroes: the phenotype Fy (a–b–). Br. J. Haematol. 1, 370–374 (1955).
Howes, R.E. et al. The global distribution of the Duffy blood group. Nat. Commun. 2, 266 (2011).
Tournamille, C., Colin, Y., Cartron, J.P. & Le Van Kim, C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat. Genet. 10, 224–228 (1995).
Peiper, S.C. et al. The Duffy antigen/receptor for chemokines (DARC) is expressed in endothelial cells of Duffy-negative individuals who lack the erythrocyte receptor. J. Exp. Med. 181, 1311–1317 (1995).
Doss, J.F. et al. A comprehensive joint analysis of the long and short RNA transcriptomes of human erythrocytes. BMC Genomics 16, 952 (2015).
Thiriot, A. et al. Differential immunostaining of DARC (ACKR1) distinguishes venular from nonvenular endothelial cells in murine tissues. BMC Biol. 15, 45 (2017).
Dawson, T.C. et al. Exaggerated response to endotoxin in mice lacking the Duffy antigen/receptor for chemokines (DARC). Blood 96, 1681–1684 (2000).
Itkin, T. et al. Distinct bone marrow blood vessels differentially regulate hematopoiesis. Nature 532, 323–328 (2016).
Morrison, S.J. & Scadden, D.T. The bone marrow niche for hematopoietic stem cells. Nature 505, 327–334 (2014).
Mendelson, A. & Frenette, P.S. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 20, 833–846 (2014).
Fossati, G., Moots, R.J., Bucknall, R.C. & Edwards, S.W. Differential role of neutrophil Fcγ receptor IIIB (CD16) in phagocytosis, bacterial killing and responses to immune complexes. Arthritis Rheum. 46, 1351–1361 (2002).
Gao, H., Henderson, A., Flynn, D.C., Landreth, K.S. & Ericson, S.G. Effects of the protein tyrosine phosphatase CD45 on FcγRIIa signaling and neutrophil function. Exp. Hematol. 28, 1062–1070 (2000).
Minten, C. et al. DARC shuttles inflammatory chemokines across the blood–brain barrier during autoimmune central nervous system inflammation. Brain 137, 1454–1469 (2014).
Martin, C. et al. Chemokines acting via CXCR2 and CXCR4 control the release of neutrophils from the bone marrow and their return following senescence. Immunity 19, 583–593 (2003).
Busch, K. et al. Fundamental properties of unperturbed hematopoiesis from stem cells in vivo. Nature 518, 542–546 (2015).
Yu, V.W. et al. Epigenetic memory underlies cell-autonomous heterogeneous behavior of hematopoietic stem cells. Cell 167, 1310–1322 e1317 (2016).
Sun, J. et al. Clonal dynamics of native hematopoiesis. Nature 514, 322–327 (2014).
Pietras, E.M. et al. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 17, 35–46 (2015).
Bandyopadhyay, S. et al. Interaction of KAI1 on tumor cells with DARC on vascular endothelium leads to metastasis suppression. Nat. Med. 12, 933–938 (2006).
Hur, J. et al. CD82 (KAI1) maintains the dormancy of long-term hematopoietic stem cells through interaction with DARC-expressing macrophages. Cell Stem Cell 18, 508–521 (2016).
Youn, B.S., Mantel, C. & Broxmeyer, H.E. Chemokines, chemokine receptors and hematopoiesis. Immunol. Rev. 177, 150–174 (2000).
Sugiyama, T., Kohara, H., Noda, M. & Nagasawa, T. Maintenance of the hematopoietic stem cell pool by CXCL12–CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25, 977–988 (2006).
Gardner, L., Patterson, A.M., Ashton, B.A., Stone, M.A. & Middleton, J. The human Duffy antigen binds selected inflammatory but not homeostatic chemokines. Biochem. Biophys. Res. Commun. 321, 306–312 (2004).
Ménard, D. et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc. Natl. Acad. Sci. USA 107, 5967–5971 (2010).
Miller, L.H., Mason, S.J., Clyde, D.F. & McGinniss, M.H. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N. Engl. J. Med. 295, 302–304 (1976).
Wright, D.E., Wagers, A.J., Gulati, A.P., Johnson, F.L. & Weissman, I.L. Physiological migration of hematopoietic stem and progenitor cells. Science 294, 1933–1936 (2001).
Seita, J. et al. Gene Expression Commons: an open platform for absolute gene expression profiling. PLoS One 7, e40321 (2012).
Faul, F., Erdfelder, E., Lang, A.-G. & Buchner, A. G*Power 3: a flexible statistical power analysis program for the social, behavioral and biomedical sciences. Behav. Res. Methods 39, 175–191 (2007).
Acknowledgements
J.D. and I.N.-B. are joint first authors, and A.T., M.C.-A., M.B., S.L.E., E.H. and K.N. contributed equally to this work. We thank M. Ulvmar, E. Ross, G. Volpe, P. Cauchy and A. Cunningham for their advice, R. Bird and S. Kissane for their assistance with cell sorting and microarray experiments, respectively, H. Vyas and P. Kelay for help with laboratory work and D. Santovito for help with statistical analysis. We thank M. Mack (University of Regensburg) and M. Uchikawa (Japanese Red Cross) for their generous gifts of antibodies specific for mouse CCR2 and human ACKR1 antibody, respectively, and J. Allen (University of Manchester) and M. Bader (Max Delbruck Center) for critical reading of the manuscript and their suggestions. A.R. is grateful to M. Tsaloumas, A. Denniston and N. Glover for the vision. Supported by Medical Research Council grant G0802838 (A.R.), a Senior Visiting Fellowship of the Center for Advanced Studies LMU, Munich (A.R.), Wellcome Trust grant WT090962MA (I.N.-B., A.R. and P.M.), Deutsche Zentrum Für Herz-Kreislauf-Forschung 86X2600229 (J.D. and C.W.), a Marie Curie Actions Intra-European Fellowship ATHEROCHEMOKINE (J.D.), Deutsche Forschungsgemeinschaft grants SFB1123/A1 (C.W.), SFB1123/Z1 (M.B. and R.T.A.M.) and INST 409/150-1 FUG (C.W. and R.T.A.M.), European Research Council grant ERC AdG °692511 (C.W.), Swiss National Science Foundation Sinergia grant CRSII3_160719 (E.H. and A.R.), a TransCard PhD fellowship in Translational Cardiovascular and Metabolic Medicine of the Helmholtz International Research School (K.N.), an ERA-EDTA short-term fellowship (K.A.) and Ministry of Economy, Industry and Competitiveness (MINECO) grant AF2015-65607-R (A.H.). The CNIC is supported by MINECO and the Pro-CNIC Foundation and is a Severo Ochoa Center of Excellence (MINECO award).
Author information
Authors and Affiliations
Contributions
A.R. conceived the study; J.D., I.N.-B., R.T.A.M., U.H.v.A., A.H., C.W. and A.R. designed the experiments; J.D., I.N.-B., A.T., M.C.-A., M.B., S.L.E., E.H., K.N., K.A. and T.R. performed the experiments and evaluated the data; J.D., I.N.-B., K.E., J.C., P.M., R.T.A.M., U.H.v.A., A.H., C.W. and A.R. interpreted the data; and A.R., J.D. and C.W. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
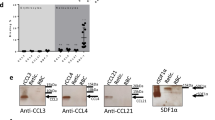
Supplementary Figure 1 Validation of the monoclonal antibody to mouse ACKR1 and expression of ACKR1 by BM hematopoietic cells.
(a to d) Comparison of immunostaining of BM cells by anti-mouse ACKR1 antibodies: new validated monoclonal (a and c, Thiriot A et al Ref. 29) and non-validated polyclonal (b and d, FAB6695P from R&D Systems used by Hur et al., Ref. 43, to report ACKR1 expression in BM macrophages). (a and b) ACKR1 immunoreactivity in nucleated erythroid cells (NECs; CD71+Ter119+), neutrophils (Neutro; CD11b+CD115–F4/80–Ly6G+), monocytes (Mono; CD11b+ CD115+F4/80–Ly6G–) and macrophages (Macro; CD11b+CD115–F4/80+Ly6G–) in BM of wild-type (WT) mice. Left, representative flow cytometry histogram plots; right, quantitative analysis. n=3. (c and d) ACKR1 immunoreactivity and negative control staining (NC) in NECs and macrophages (Macro) in BM of WT (blue) and ACKR1-deficient (KO, red) mice. Left, representative flow cytometry histogram plots; right, quantitative analysis. n=3. BM immunostaining using validated monoclonal antibody (a and c) show that ACKR1 is expressed in the BM by erythroid cells only but not by macrophages. Polyclonal non-validated antibody failed to immunoreact with NECs but at high concentration stained BM macrophages, (b) however non-specifically as (d) to the same extent in WT and KO mice. All data are Mean±SEM. One-Way ANOVA (a and b). Two-Way ANOVA (c and d). *P<0.001. (e) Relative expression of ACKR1 in hematopoietic and stromal populations as determined by microarray analysis based on the data in the BM cells transcriptome database (https://gexc.riken.jp/models). Microarray dataset of adult definitive murine erythroblasts were retrieved from ArrayExpress (https://www.ebi.ac.uk/arrayexpress; E-MTAB-1035) and implemented in Gene Expression Commons database (link to this model https://gexc.riken.jp/models/1649). Proerythroblasts (ProE), early normoblasts (BasoE), late normoblasts (PolyE), reticulocytes (Ret) and Endothelial cells (BM EC) in BM express ACKR1 whereas BM macrophages (BM Macro) do not.
Supplementary Figure 2 ACKR1 is expressed by endothelial cells and nucleated erythroid cells in the BM but not by other hematopoietic cells.
(a) Immunofluorescence micrographs of wild-type (WT) and ACKR1-deficient (KO) BM stained with anti-ACKR1 (red), anti-CD31 (endothelial cells, blue), anti-Ter119 (erythroid cells, yellow) antibodies and DAPI (nuclei, turquoise). (b) ACKR1 expression (MFI) on HSPCs, megakaryocyte progenitors (MkP) and leukocytes (Leu) as compared to the subpopulations of erythroid cells at different stages of development (I-VI) in WT (red) and KO (blue) BM (Mean±SEM; n=3). Scale bar, 30 μm (c) Human ACKR1 mRNA BM expression data retrieved from DMAP (http://portals.broadinstitute.org/dmap/home). Subpopulations of erythroid cells (ERY1-4) were defined as follows: CD71hi GlyA–(Ery1), CD71hi GlyA+(Ery2), CD71lo GlyA+(Ery3) and CD71–GlyA+(Ery4). Mean±SEM, One-Way ANOVA; *P < 0.001.
Supplementary Figure 3 ACKR1 deficiency does not affect erythroid parameters in BM and blood.
(a) BM cellularity in in WT (blue) and ACKR1-deficient (KO; red) mice (Mean±SEM; n=9). (b) Percentage of proerythroblasts (I), early normoblasts (II), intermediate normoblasts (III), late normoblasts (IV), reticulocytes (V) and mature red cells (VI) in BM of WT and KO mice (Mean±SEM; n=3). (c) Erythrocyte parameters in blood. RBC: Red blood cell counts; HCT: Haematocrit; Hb: haemoglobin. (Mean±SEM; n=9).
Supplementary Figure 4 Flow cytometry analysis of HSPCs, CD48+ subpopulations of LSK cells and CLP cells.
(a) Gating strategy to identify HSPC populations. LSK, defined as Lin–Sca-1+c-Kit+ were subdivided into LSK CD48+ and LSK CD48+. Myeloid progenitor cells (MPC) were defined as Lin–Sca-1–c-Kit+. (b) Frequency of LSK CD48+ cells in in WT (blue) and ACKR1-deficient (KO, red) BM. (c) Relative distribution of their MPP2, and lineage-restricted progenitor (LRP) 1 and LRP2 sub-populations. Representative dot plots, left; quantitative analysis, right. (d) Frequency of MPP2, LRP1 and LRP2 in the BM of WT and KO mice. (e) Frequency of common lymphoid progenitors (CLP) defined as Lin–Sca-1loc-Kitlo (G1 gate in a) and IL7Rɑ+Ftl3+. Representative dot plots, left; quantitative analysis, right. (b-d) n=12 in four independent experiments. (f) n=5 in two independent experiments. All data show Mean±SEM. Two-tailed Student’s t-test; ***p < 0.001.
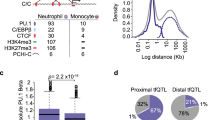
Supplementary Figure 5 Gene expression in LSKs and GMPs from WT and ACKR1-deficient mice.
(a) Microarray heatmap of genes expressed in LSKs and GMPs from wild-type (WT) and ACKR1-deficient (KO) BMs. Each row represents a gene and columns show individual cell populations, in duplicates. Two clusters defined by hierarchical clustering analysis reflect specific gene expression in LSK and GMP. The levels of expression of specific LSK and GMP enriched genes were similar in WT and KO BM. (b) Left, blue: Comparative gene expression in neutrophils (PMNs) and GMPs from Immgen dataset (www.immgen.org). The expression of the 444 genes was higher in PMNs vs. GMPs on average >10 times. Three genes with the highest increase, Ltf, Ly6G and Mmp8, were expressed ca. 100 times more in PMN, than in GMP. Right, red: comparative gene expression in GMPs from WT vs. KO BM expressed as fold change. The level of expression of the overwhelming majority of genes was the same in GMPs from WT and KO BMs (mean fold change 1) but some were expressed orders of magnitude higher in KO GMPs. The transcripts for three most overexpressed genes Rentlg, Camp and Ngp, were in excess of 1000 times higher in KO vs. WT GMPs. These genes were not highly upregulated during the differentiation of GMPs towards PMNs (left panel, blue). This excluded a potential contamination of the GMP with PMNs and was consistent with the induction of a specific transcriptional program of a subset of neutrophil specific genes (also see Fig. 3c) in GMPs rather than only their conventional differentiation to PMNs. (c) Microarray heatmap from Fig. 3a with the list of genes. (d) Microarray heatmap from Fig. 3b with the list of genes.
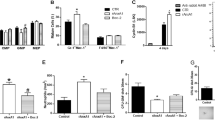
Supplementary Figure 6 BM and blood cell parameters in reciprocal irradiation BM chimeric and parabiotic wild-type and ACKR1-deficient mice.
(a) Schematic representation of experimental groups of irradiation BM chimeric mice. (b,c) Confirmation of successful BM chimerism by flow cytometric measurement of ACKR1 expression on erythroid cells in BM and in blood. (b) Quantitative analysis of ACKR1 expression on subpopulations of erythroid cells (I-VI) in BM. (c) Percentages of ACKR1+and ACKR1– RBC in peripheral blood. (d) Relative distribution of MPP2, LRP1 and LRP2 subpopulation of LSK CD48+ in irradiation BM chimeric mice: wild-type (WT) BM cells reconstituted into WT mice (blue), WT BM cells reconstituted into ACKR1-deficient (KO) mice (light blue), KO BM cells reconstituted into KO mice (red) and KO BM cells reconstituted into WT mice (pink); right, representative dot plots; left, quantitative analysis. (e) Schematic representation of experimental groups in parabiosis experiments. (f) Confirmation of shared circulation in parabiosis by measurement of plasma CCL2 levels in individual naïve and parabiotic mice. WT with WT parabionts (blue), KO with KO parabionts (red), WT parabiont in WT with KO pair (purple) and KO parabiont in WT with KO pair (orange) (g) Relative distribution of MPP2, LRP1 and LRP2 subpopulation of LSK CD48+, right, representative dot plots; left, quantitative analysis. All data are n=4 from two independent experiments, Mean±SEM. One-way ANOVA; *P < 0.05, **P < 0.01 and ***P < 0.001.
Supplementary Figure 7 Interactions of HSCs and nucleated erythroid cells in the BM.
(a) Representative flow cytometry histograms showing staining by antibodies against CD48, Lineage (Lin; B220, CD3, CD11b, Gr1), CD150 and CD71 used for identification of HSCs and nucleated erythroid cells (NECs). The HSCs (LSK CD150+ CD48–) are distinguished as Lin– CD150+CD48–CD71–; the NECs (CD71+ Ter119+) are CD71+Lin–CD150–CD48–; the MPCs and CD48+LSK subset are Lin–CD48+ CD150–CD71– and the lineage cells are Lin+CD48+. (b) Application of the above combination of antibodies to the two-photon microscopy of the whole-mounted femur. HSCs (Lin–CD150+CD71–; green) and NECs (Lin–CD150– CD71+; red) appear clearly distinct. HSCs (in the upper rows of magnification insets, green; in lower rows of insets, only outlined in white) did not stain for lineage or CD71. Scale bars, 50 μm and 40 μm (c) Representative two photon microscopy images of the whole-mounted femurs from ACKR1-deficient mice used for the measurements of the distance between NECs and HSCs. (d) Quantitative analysis of HSC, MMP1, MPP2 and LRP populations interacting with NECs assessed by flow cytometry. Wild-type (WT, blue) and ACKR1-deficient (KO, red). Two-tailed Student’s t-test, (n=3). All numeric data are Mean±SEM. ***P < 0.001.
Supplementary Figure 8 Effect of ACKR1 on the expression of cell surface markers and cell numbers in BM and blood.
(a) CD62L, CD11a, CD16/32, CD45 and CD11b expression in BM neutrophils (CD11b+Ly6G+) from wild-type (WT, blue) and ACKR1-deficient (KO, red) mice; NC (white), negative control staining. Top, representative flow cytometry histograms; bottom, quantitative analysis. (b) Representative flow cytometry histograms of staining blood neutrophils (CD11b+Ly6G+) from WT and KO mice with antibodies against CD62L and CD11a and (c) antibodies against CXCR2, CCR1, CCR2, CCR3 and CCR5. (d) Representative flow cytometry histograms of CXCR1, CXCR2 and CCR2 antibodies staining of blood neutrophils from Duffy-positive and Duffy-negative individuals, grey and black histograms, respectively, negative control staining, NC (white). (e) PMN (CD11b+Ly6G+CD115–), monocyte (CD11b+Ly6G–CD115+) and B lineage cell (B220+CD19+) counts in BM of WT and ACKR1-deficient mice (n=6). (f) Monocyte (CD45+CD11b+Ly6G– CD115+) and lymphoid cell (CD45+ and B220+ or CD3+) counts in blood of WT and ACKR1-deficient mice (n=6). (g) Platelet counts in blood of WT and ACKR1-deficient mice (n=16). (h) Expression of CD16/32 on blood neutrophils (CD11b+Ly6G+CD115–) in reciprocal BM chimeric mice; WT BM cells reconstituted into WT mice (blue), WT BM cells reconstituted into KO mice (light blue), KO BM cells reconstituted into KO mice (red) and KO BM cells reconstituted into WT mice (pink); n=4 from two independent experiments. (i) Representative immunofluorescence micrograph of human spleen stained with monoclonal anti-ACKR1 (Fy6, red) antibody. All data show Mean±SEM. (a and e-g) two-tailed Student’s t-test; *P < 0.05, **P < 0.01. (h) one-way ANOVA; **P < 0.01.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Duchene, J., Novitzky-Basso, I., Thiriot, A. et al. Atypical chemokine receptor 1 on nucleated erythroid cells regulates hematopoiesis. Nat Immunol 18, 753–761 (2017). https://doi.org/10.1038/ni.3763
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3763
This article is cited by
-
The chemokines CXCL8 and CXCL12: molecular and functional properties, role in disease and efforts towards pharmacological intervention
Cellular & Molecular Immunology (2023)
-
Niche derived netrin-1 regulates hematopoietic stem cell dormancy via its receptor neogenin-1
Nature Communications (2021)
-
Differential interaction between DARC and SDF-1 on erythrocytes and their precursors
Scientific Reports (2019)
-
A genome-wide association study in individuals of African ancestry reveals the importance of the Duffy-null genotype in the assessment of clozapine-related neutropenia
Molecular Psychiatry (2019)
-
Dual role for atypical chemokine receptor 1 in myeloid cell hematopoiesis and distribution
Cellular & Molecular Immunology (2018)