Abstract
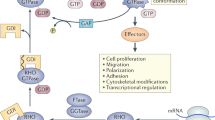
RASGRP1 is an important guanine nucleotide exchange factor and activator of the RAS-MAPK pathway following T cell antigen receptor (TCR) signaling. The consequences of RASGRP1 mutations in humans are unknown. In a patient with recurrent bacterial and viral infections, born to healthy consanguineous parents, we used homozygosity mapping and exome sequencing to identify a biallelic stop-gain variant in RASGRP1. This variant segregated perfectly with the disease and has not been reported in genetic databases. RASGRP1 deficiency was associated in T cells and B cells with decreased phosphorylation of the extracellular-signal-regulated serine kinase ERK, which was restored following expression of wild-type RASGRP1. RASGRP1 deficiency also resulted in defective proliferation, activation and motility of T cells and B cells. RASGRP1-deficient natural killer (NK) cells exhibited impaired cytotoxicity with defective granule convergence and actin accumulation. Interaction proteomics identified the dynein light chain DYNLL1 as interacting with RASGRP1, which links RASGRP1 to cytoskeletal dynamics. RASGRP1-deficient cells showed decreased activation of the GTPase RhoA. Treatment with lenalidomide increased RhoA activity and reversed the migration and activation defects of RASGRP1-deficient lymphocytes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Roose, J. & Weiss, A. T cells: getting a GRP on Ras. Nat. Immunol. 1, 275–276 (2000).
Stone, J.C. Regulation and function of the RasGRP family of Ras activators in blood cells. Genes Cancer 2, 320–334 (2011).
Downward, J., Graves, J.D., Warne, P.H., Rayter, S. & Cantrell, D.A. Stimulation of p21ras upon T-cell activation. Nature 346, 719–723 (1990).
Kremer, K.N., Kumar, A. & Hedin, K.E. G alpha i2 and ZAP-70 mediate RasGRP1 membrane localization and activation of SDF-1-induced T cell functions. J. Immunol. 187, 3177–3185 (2011).
Dower, N.A. et al. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat. Immunol. 1, 317–321 (2000).
Khanna, R. & Burrows, S.R. Human immunology: a case for the ascent of non-furry immunology. Immunol. Cell Biol. 89, 330–331 (2011).
Mestas, J. & Hughes, C.C. Of mice and not men: differences between mouse and human immunology. J. Immunol. 172, 2731–2738 (2004).
Waterston, R.H. et al. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002).
Notarangelo, L.D. Functional T cell immunodeficiencies (with T cells present). Annu. Rev. Immunol. 31, 195–225 (2013).
Notarangelo, L.D. Partial defects of T-cell development associated with poor T-cell function. J. Allergy Clin. Immunol. 131, 1297–1305 (2013).
Picard, C. et al. Primary immunodeficiency diseases: an update on the classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J. Clin. Immunol. 35, 696–726 (2015).
Bousfiha, A. et al. The 2015 IUIS phenotypic classification for primary immunodeficiencies. J. Clin. Immunol. 35, 727–738 (2015).
Espinós, C. et al. Mutations in the urocanase gene UROC1 are associated with urocanic aciduria. J. Med. Genet. 46, 407–411 (2009).
Chen, Y. et al. Differential requirement of RasGRP1 for γδ T cell development and activation. J. Immunol. 189, 61–71 (2012).
Warnecke, N. et al. TCR-mediated Erk activation does not depend on Sos and Grb2 in peripheral human T cells. EMBO Rep. 13, 386–391 (2012).
Jun, J.E., Rubio, I. & Roose, J.P. Regulation of ras exchange factors and cellular localization of ras activation by lipid messengers in T cells. Front. Immunol. 4, 239 (2013).
Depeille, P. et al. RasGRP1 opposes proliferative EGFR-SOS1-Ras signals and restricts intestinal epithelial cell growth. Nat. Cell Biol. 17, 804–815 (2015).
Shen, S. et al. Critical roles of RasGRP1 for invariant NKT cell development. J. Immunol. 187, 4467–4473 (2011).
Montoya, C.J. et al. Characterization of human invariant natural killer T subsets in health and disease using a novel invariant natural killer T cell-clonotypic monoclonal antibody, 6B11. Immunology 122, 1–14 (2007).
Priatel, J.J. et al. Chronic immunodeficiency in mice lacking RasGRP1 results in CD4 T cell immune activation and exhaustion. J. Immunol. 179, 2143–2152 (2007).
Ma, C.S. & Deenick, E.K. Human T follicular helper (Tfh) cells and disease. Immunol. Cell Biol. 92, 64–71 (2014).
Bartlett, A., Buhlmann, J.E., Stone, J., Lim, B. & Barrington, R.A. Multiple checkpoint breach of B cell tolerance in Rasgrp1-deficient mice. J. Immunol. 191, 3605–3613 (2013).
Coughlin, J.J., Stang, S.L., Dower, N.A. & Stone, J.C. RasGRP1 and RasGRP3 regulate B cell proliferation by facilitating B cell receptor-Ras signaling. J. Immunol. 175, 7179–7184 (2005).
Sun, C. et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat. Genet. 48, 323–330 (2016).
Tangye, S.G., Ferguson, A., Avery, D.T., Ma, C.S. & Hodgkin, P.D. Isotype switching by human B cells is division-associated and regulated by cytokines. J. Immunol. 169, 4298–4306 (2002).
Thien, M. et al. Excess BAFF rescues self-reactive B cells from peripheral deletion and allows them to enter forbidden follicular and marginal zone niches. Immunity 20, 785–798 (2004).
Perussia, B., Chen, Y. & Loza, M.J. Peripheral NK cell phenotypes: multiple changing of faces of an adapting, developing cell. Mol. Immunol. 42, 385–395 (2005).
Navarro, M.N. & Cantrell, D.A. Serine-threonine kinases in TCR signaling. Nat. Immunol. 15, 808–814 (2014).
Okamura, S.M., Oki-Idouchi, C.E. & Lorenzo, P.S. The exchange factor and diacylglycerol receptor RasGRP3 interacts with dynein light chain 1 through its C-terminal domain. J. Biol. Chem. 281, 36132–36139 (2006).
Rodríguez-Crespo, I. et al. Identification of novel cellular proteins that bind to the LC8 dynein light chain using a pepscan technique. FEBS Lett. 503, 135–141 (2001).
Huse, M. Microtubule-organizing center polarity and the immunological synapse: protein kinase C and beyond. Front. Immunol. 3, 235 (2012).
Mentlik, A.N., Sanborn, K.B., Holzbaur, E.L. & Orange, J.S. Rapid lytic granule convergence to the MTOC in natural killer cells is dependent on dynein but not cytolytic commitment. Mol. Biol. Cell 21, 2241–2256 (2010).
Roose, J.P., Mollenauer, M., Gupta, V.A., Stone, J. & Weiss, A. A diacylglycerol-protein kinase C-RasGRP1 pathway directs Ras activation upon antigen receptor stimulation of T cells. Mol. Cell. Biol. 25, 4426–4441 (2005).
Li, R. & Gundersen, G.G. Beyond polymer polarity: how the cytoskeleton builds a polarized cell. Nat. Rev. Mol. Cell Biol. 9, 860–873 (2008).
Liu, Y., Zhu, M., Nishida, K., Hirano, T. & Zhang, W. An essential role for RasGRP1 in mast cell function and IgE-mediated allergic response. J. Exp. Med. 204, 93–103 (2007).
Ramsay, A.G. et al. Chronic lymphocytic leukemia cells induce defective LFA-1-directed T-cell motility by altering Rho GTPase signaling that is reversible with lenalidomide. Blood 121, 2704–2714 (2013).
Richardson, P.G. et al. Lenalidomide, bortezomib, and dexamethasone combination therapy in patients with newly diagnosed multiple myeloma. Blood 116, 679–686 (2010).
Krönke, J. et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 (2014).
Gandhi, A.K. et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.). Br. J. Haematol. 164, 811–821 (2014).
Casanova, J.L., Conley, M.E., Seligman, S.J., Abel, L. & Notarangelo, L.D. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J. Exp. Med. 211, 2137–2149 (2014).
Fuller, D.M. et al. Regulation of RasGRP1 function in T cell development and activation by its unique tail domain. PLoS One 7, e38796 (2012).
Vivier, E., Tomasello, E., Baratin, M., Walzer, T. & Ugolini, S. Functions of natural killer cells. Nat. Immunol. 9, 503–510 (2008).
Thrasher, A.J. & Burns, S.O. WASP: a key immunological multitasker. Nat. Rev. Immunol. 10, 182–192 (2010).
Lanzi, G. et al. A novel primary human immunodeficiency due to deficiency in the WASP-interacting protein WIP. J. Exp. Med. 209, 29–34 (2012).
McGhee, S.A. & Chatila, T.A. DOCK8 immune deficiency as a model for primary cytoskeletal dysfunction. Dis. Markers 29, 151–156 (2010).
Liu, X., Kapoor, T.M., Chen, J.K. & Huse, M. Diacylglycerol promotes centrosome polarization in T cells via reciprocal localization of dynein and myosin II. Proc. Natl. Acad. Sci. USA 110, 11976–11981 (2013).
Schroeder, C.M., Ostrem, J.M., Hertz, N.T. & Vale, R.D. A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. eLife 3, e03351 (2014).
Jayappa, K.D. et al. Human immunodeficiency virus type 1 employs the cellular dynein light chain 1 protein for reverse transcription through interaction with its integrase protein. J. Virol. 89, 3497–3511 (2015).
Zenz, T. Exhausting T cells in CLL. Blood 121, 1485–1486 (2013).
Salzer, E. et al. Combined immunodeficiency with life-threatening EBV-associated lymphoproliferative disorder in patients lacking functional CD27. Haematologica 98, 473–478 (2013).
Willmann, K.L. et al. Biallelic loss-of-function mutation in NIK causes a primary immunodeficiency with multifaceted aberrant lymphoid immunity. Nat. Commun. 5, 5360 (2014).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Dupré, L. et al. Efficacy of gene therapy for Wiskott-Aldrich syndrome using a WAS promoter/cDNA-containing lentiviral vector and nonlethal irradiation. Hum. Gene Ther. 17, 303–313 (2006).
Dupré, L. et al. Lentiviral vector-mediated gene transfer in T cells from Wiskott-Aldrich syndrome patients leads to functional correction. Mol. Ther. 10, 903–915 (2004).
Pichlmair, A. et al. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 487, 486–490 (2012).
Glatter, T., Wepf, A., Aebersold, R. & Gstaiger, M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 5, 237 (2009).
Dow, L.E. et al. A pipeline for the generation of shRNA transgenic mice. Nat. Protoc. 7, 374–393 (2012).
Zuber, J. et al. An integrated approach to dissecting oncogene addiction implicates a Myb-coordinated self-renewal program as essential for leukemia maintenance. Genes Dev. 25, 1628–1640 (2011).
Ran, F.A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Sanjana, N.E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Brinkman, E.K., Chen, T., Amendola, M. & van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42, e168 (2014).
Boztug, K. et al. JAGN1 deficiency causes aberrant myeloid cell homeostasis and congenital neutropenia. Nat. Genet. 46, 1021–1027 (2014).
Mellacheruvu, D. et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 (2013).
Vasconcelos, Z. et al. Individual human cytotoxic T lymphocytes exhibit intraclonal heterogeneity during sustained killing. Cell Rep. 11, 1474–1485 (2015).
Huppa, J.B. et al. TCR-peptide-MHC interactions in situ show accelerated kinetics and increased affinity. Nature 463, 963–967 (2010).
Fairhead, M., Krndija, D., Lowe, E.D. & Howarth, M. Plug-and-play pairing via defined divalent streptavidins. J. Mol. Biol. 426, 199–214 (2014).
Howarth, M. et al. A monovalent streptavidin with a single femtomolar biotin binding site. Nat. Methods 3, 267–273 (2006).
Banerjee, P.P. & Orange, J.S. Quantitative measurement of F-actin accumulation at the NK cell immunological synapse. J. Immunol. Methods 355, 1–13 (2010).
Trifari, S. et al. Defective Th1 cytokine gene transcription in CD4+ and CD8+ T cells from Wiskott-Aldrich syndrome patients. J. Immunol. 177, 7451–7461 (2006).
Acknowledgements
We thank the patient and his family for participating in this study; B. Fleckenstein and M. Schmidt for generating and providing patient-derived T cell lines; and G. Superti-Furga, J. Bigenzahn for providing the inducible protein expression system used for Jurkat T cells and together with N. Serwas, C.D. Conde, A. Kalinichenko, K. Ackerman and R. Martins for critically reviewing the manuscript and providing comments. The research leading to these results was funded by the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement 310857 (K.B.), the Vienna Science and Technology Fund (WWTF) through project LS14-031 (J.B.H. and K.B.), an unrestricted research grant from Celgene Austria (U.J.), the National Institutes of Health (R01AI067946 to J.S.O.), Boehringer Ingelheim Fonds (R.P.), the Austrian Science Fund (FWF): Project M1809-B19 (K.L.W.), and the French Agence Nationale de la Recherche (ANR-13-BSV1-0031 to L.D.).
Author information
Authors and Affiliations
Contributions
E.S. performed most of the experiments, analyzed data, interpreted results, and, together with K.B., wrote the initial draft and revised version of the manuscript. D.C., I.T. and Ö.S. cared for the patient, and provided and interpreted clinical and immunological data. M.H. and E.S. performed migration and Lifeact experiments. M.S. provided critical input to the content of the manuscript. E.M.M., P.S., M.M., P.P.B., H.T.H. and J.S.O. performed and interpreted NK-cell immunological synapse experiments and detailed flow-cytometry-based NK-cell immunophenotyping. S.A.B. and E.S. identified the RASGRP1 mutation and performed initial experiments. R.P. and J.B.H. performed lipid bilayer calcium-flux experiments, and analyzed and interpreted data. L.P. generated an untransformed CD8+ T cell line from the patient and performed the CD8+ T cell cytotoxic assays. H.S. and V.S. performed proliferation analyses together with E.S. and provided critical input. Ö.Y.P. and L.D. performed the immunofluorescence experiments to quantify RhoA activation. K.L.W., W.G. and I.B. performed experiments and provided critical input. F.M., J.G.G. and U.J. helped with migration analyses. K.L.B. performed mass spectrometry analyses. W.F.P. and G.J.Z. performed thymidine incorporation assays, chromium release assays and analyses of autoantibody titers. K.B. conceived of and coordinated the study, provided laboratory resources, interpreted data, supervised E.S., W.G., Ö.Y.P., I.B., S.B. and K.W., wrote the manuscript together with E.S. and took overall responsibility for the study. All of the authors provided critical input and agreed to this publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
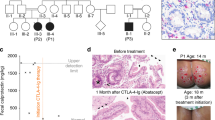
Supplementary Figure 1 Histology of lymphoma and segregation of the mutation with the disease phenotype.
(a) Histology of adenoid biopsy showing low grade B-cell lymphoma compatible with marginal zone lymphoma. Top left to right: Hematoxilin/Eosin staining, CD20 staining, CD3 staining. Bottom left to right: EBER, CD23, Ki67. The described findings suggest a low grade EBV-related B-cell lymphoma developed likely associated with the underlying immunodeficiency. (b) Segregation of the detected mutation among the core family (circle - female; square - male; line - dead; black filling - affected index patient).
Supplementary Figure 2 Functional characterization of T cells.
(a) Invariant natural killer T (iNKT) cells. A prominent TCRvα24-expressing cell population was detectable in patient peripheral blood, however these cells expressed CD8 and CD45RA surface markers, suggesting that they belong to oligoclonally expanded exhausted memory CD8 T cells. HD – healthy donor; SC – Shipment control. (b) Proliferative response of T cells determined by [3H]-thymidine incorporation assay after stimulation with various stimuli after 3 days (OKT3, anti-CD3 antibody (clone OKT3)); PMA, phorbol 12-myristate 13-acetate; SEA: Staphylococcal enterotoxin A; SEB: Staphylococcal enterotoxin B; PHA: phytohaemagglutinin). (c) Proliferation of Jurkat T cells upon shRNA-mediated knockdown of RASGRP1. shRNA against Renilla luciferase was used as negative control. Cells were labeled with violet proliferation dye and analyzed by flow cytometry over the period of 5 days. (d) TCRVb spectratyping of patient and control T cells indicating oligoclonality of the TCR repertoire of the patient. (e) Calcium flux of Jurkat T cells upon inducible shRNA-mediated knockdown of RASGRP-1. shRNA against Renilla luciferase was used as negative control. (f) Fluorescence microscopy of Ca2+-flux of one representative cell is displayed below the graph. Scale bar represents 10 μm. HD–healthy donor; DIC–differential interference contrast. (g) ICAM Ring formation and cSMAC exclusion of hTERT immortalized patient and control T cell lines on a lipid bilayer following OKT3 stimulation. (h) Cropped immunoblot showing downstream TCR signaling in expanded patient and shipment T cells upon CD3/CD28 stimulation. Cells were starved and restimulated with anti-CD3/anti-CD28 antibodies and subsequently analyzed for ERK1/2 phosphorylation. Data is representative of three (h), two (a,c,d,g) or one (b,f,e) independent experiment.
Supplementary Figure 4 NK-cell studies.
(a) Flow cytometric analysis of intracellular accumulation of IL-5, IL13 and IFNγ in patient and control NK cells. (b) Immune synapse formation in primary patient NK cells (c) Co-immunoprecipitation of Strep-HA tagged RASGRP1 with endogenous dynein light chain 1 (DYNLL1) (RASGRP1wt) wildtype Strep-HA-tagged RASGRP1 and (RASGRP1mut) mutant Strep-HA-tagged RASGRP1. Strep-HA-tagged GFP was used as negative control. (d) Video microscopy of granule convergence in CRISPR-edited NK-92 cells lines HD, healthy donor. (d) Video microscopy of control or sgRASGRP1 NK-92 cells for granule convergence (Granules-red, Microtubuli-green). Data are representative of two biological replicates (a-d). (e) Gating strategy of FACS plots in Figure 4.
Supplementary Figure 5 T-cell activation using CXCL12.
(a-c) Immunofluorescence (a) and quantification (b,c) of patient and healthy donor expanded CD8 T cells following CXCL12 stimulation either stained for total RhoA (green) or active RhoA (pink) and DAPI (blue). Data is representative of two (a-c) independent experiments.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1–8 (PDF 2516 kb)
Rights and permissions
About this article
Cite this article
Salzer, E., Cagdas, D., Hons, M. et al. RASGRP1 deficiency causes immunodeficiency with impaired cytoskeletal dynamics. Nat Immunol 17, 1352–1360 (2016). https://doi.org/10.1038/ni.3575
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.3575
This article is cited by
-
Unraveling the dynamic mechanisms of natural killer cells in viral infections: insights and implications
Virology Journal (2024)
-
B-cells absence in patients diagnosed as inborn errors of immunity: a registry-based study
Immunogenetics (2024)
-
Phosphorylation of RasGRP1 by Shc3 prevents RasGRP1 degradation and contributes to Ras/c-Jun activation in hepatocellular carcinoma
Molecular and Cellular Biochemistry (2023)
-
Genetic regulation and variation of expression of miRNA and mRNA transcripts in fetal muscle tissue in the context of sex, dam and variable fetal weight
Biology of Sex Differences (2022)
-
Novel PGM3 mutation in two siblings with combined immunodeficiency and childhood bullous pemphigoid: a case report and review of the literature
Allergy, Asthma & Clinical Immunology (2022)