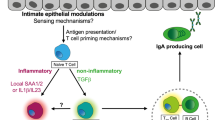
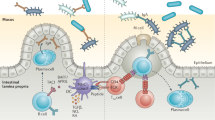
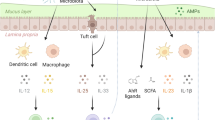
Abstract
Pattern-recognition receptors (PRRs) are traditionally known to sense microbial molecules during infection to initiate inflammatory responses. However, ligands for PRRs are not exclusive to pathogens and are abundantly produced by the resident microbiota during normal colonization. Mechanism(s) that underlie this paradox have remained unclear. Recent studies reveal that gut bacterial ligands from the microbiota signal through PRRs to promote development of host tissue and the immune system, and protection from disease. Evidence from both invertebrate and vertebrate models reveals that innate immune receptors are required to promote long-term colonization by the microbiota. This emerging perspective challenges current models in immunology and suggests that PRRs may have evolved, in part, to mediate the bidirectional cross-talk between microbial symbionts and their hosts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Janeway, C.A. Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
Beutler, B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature 430, 257–263 (2004).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
Meylan, E., Tschopp, J. & Karin, M. Intracellular pattern recognition receptors in the host response. Nature 442, 39–44 (2006).
Kawai, T. & Akira, S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–384 (2010).
Backhed, F., Ley, R.E., Sonnenburg, J.L., Peterson, D.A. & Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 307, 1915–1920 (2005).
Dethlefsen, L., McFall-Ngai, M. & Relman, D.A. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449, 811–818 (2007).
Lee, Y.K. & Mazmanian, S.K. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330, 1768–1773 (2010).
Hooper, L.V. et al. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291, 881–884 (2001).
Nyholm, S.V. & Graf, J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Microbiol. 10, 815–827 (2012).
Mackey, D. & McFall, A.J. MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol. Microbiol. 61, 1365–1371 (2006).
Round, J.L. & Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9, 313–323 (2009).
Kubinak, J.L. & Round, J.L. Toll-like receptors promote mutually beneficial commensal-host interactions. PLoS Pathog. 8, e1002785 (2012).
Matzinger, P. Tolerance, danger, and the extended family. Annu. Rev. Immunol. 12, 991–1045 (1994).
Vance, R.E., Isberg, R.R. & Portnoy, D.A. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6, 10–21 (2009).
Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J.M. & Hoffmann, J.A. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 (1996).
Werner, T. et al. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97, 13772–13777 (2000).
Michel, T., Reichhart, J.M., Hoffmann, J.A. & Royet, J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414, 756–759 (2001).
Gottar, M. et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416, 640–644 (2002).
Gay, N.J. & Keith, F.J. Drosophila Toll and IL-1 receptor. Nature 351, 355–356 (1991).
Levashina, E.A. et al. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science 285, 1917–1919 (1999).
Choe, K.M., Werner, T., Stoven, S., Hultmark, D. & Anderson, K.V. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296, 359–362 (2002).
Ramet, M., Manfruelli, P., Pearson, A., Mathey-Prevot, B. & Ezekowitz, R.A. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416, 644–648 (2002).
Ryu, J.H. et al. The homeobox gene Caudal regulates constitutive local expression of antimicrobial peptide genes in Drosophila epithelia. Mol. Cell. Biol. 24, 172–185 (2004).
Kleino, A. et al. Pirk is a negative regulator of the Drosophila Imd pathway. J. Immunol. 180, 5413–5422 (2008).
Lhocine, N. et al. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe 4, 147–158 (2008).
Bischoff, V. et al. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2, e14 (2006).
Paredes, J.C., Welchman, D.P., Poidevin, M. & Lemaitre, B. Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35, 770–779 (2011).
Franzenburg, S. et al. Bacterial colonization of Hydra hatchlings follows a robust temporal pattern. ISME J. 7, 781–790 (2013).
Bosch, T.C. et al. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Dev. Comp. Immunol. 33, 559–569 (2009).
Kobe, B. & Deisenhofer, J. Proteins with leucine-rich repeats. Curr. Opin. Struct. Biol. 5, 409–416 (1995).
Fraune, S. et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc. Natl. Acad. Sci. USA 107, 18067–18072 (2010).
Franzenburg, S. et al. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc. Natl. Acad. Sci. USA 109, 19374–19379 (2012).
Nyholm, S.V. & McFall-Ngai, M.J. The winnowing: establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol. 2, 632–642 (2004).
McFall-Ngai, M., Nyholm, S.V. & Castillo, M.G. The role of the immune system in the initiation and persistence of the Euprymna scolopes–Vibrio fischeri symbiosis. Semin. Immunol. 22, 48–53 (2010).
McFall-Ngai, M., Heath-Heckman, E.A., Gillette, A.A., Peyer, S.M. & Harvie, E.A. The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin. Immunol. 24, 3–8 (2012).
McFall-Ngai, M.J. & Ruby, E.G. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254, 1491–1494 (1991).
Koropatnick, T.A. et al. Microbial factor-mediated development in a host-bacterial mutualism. Science 306, 1186–1188 (2004).
Goodson, M.S. et al. Euprymna scolopes–Vibrio fischeri light organ symbiosis. Appl. Environ. Microbiol. 71, 6934–6946 (2005).
Foster, J.S., Apicella, M.A. & McFall-Ngai, M.J. Vibrio fischeri lipopolysaccharide induces developmental apoptosis, but not complete morphogenesis, of the Euprymna scolopes symbiotic light organ. Dev. Biol. 226, 242–254 (2000).
Wang, J. & Aksoy, S. PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse's offspring. Proc. Natl. Acad. Sci. USA 109, 10552–10557 (2012).
Collins, A.J., Schleicher, T.R., Rader, B.A. & Nyholm, S.V. Understanding the role of host hemocytes in a squid/vibrio symbiosis using transcriptomics and proteomics. Front. Immunol. 3, 91 (2012).
Troll, J.V. et al. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell. Microbiol. 11, 1114–1127 (2009).
Troll, J.V. et al. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ. Microbiol. 12, 2190–2203 (2010).
Rawls, J.F., Samuel, B.S. & Gordon, J.I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. USA 101, 4596–4601 (2004).
Nasevicius, A. & Ekker, S.C. Effective targeted gene 'knockdown' in zebrafish. Nat. Genet. 26, 216–220 (2000).
De Rienzo, G., Gutzman, J.H. & Sive, H. Efficient shRNA-mediated inhibition of gene expression in zebrafish. Zebrafish 9, 97–107 (2012).
Pham, L.N., Kanther, M., Semova, I. & Rawls, J.F. Methods for generating and colonizing gnotobiotic zebrafish. Nat. Protoc. 3, 1862–1875 (2008).
Stein, C., Caccamo, M., Laird, G. & Leptin, M. Conservation and divergence of gene families encoding components of innate immune response systems in zebrafish. Genome Biol. 8, R251 (2007).
Bates, J.M., Akerlund, J., Mittge, E. & Guillemin, K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe 2, 371–382 (2007).
Meijer, A.H. et al. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol. Immunol. 40, 773–783 (2004).
Sullivan, C. et al. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J. Immunol. 183, 5896–5908 (2009).
Sepulcre, M.P. et al. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J. Immunol. 182, 1836–1845 (2009).
Rader, B.A., Kremer, N., Apicella, M.A., Goldman, W.E. & McFall-Ngai, M.J. Modulation of symbiont lipid A signaling by host alkaline phosphatases in the squid-vibrio symbiosis. MBio 3, e00093–12 (2012).
Goldberg, R.F. et al. Intestinal alkaline phosphatase is a gut mucosal defense factor maintained by enteral nutrition. Proc. Natl. Acad. Sci. USA 105, 3551–3556 (2008).
Malo, M.S. et al. Intestinal alkaline phosphatase preserves the normal homeostasis of gut microbiota. Gut. 59, 1476–1484 (2010).
Johansson, M.E., Larsson, J.M. & Hansson, G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. USA 108 (suppl. 1), 4659–4665 (2011).
Johansson, M.E., Sjovall, H. & Hansson, G.C. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. advance online publication, doi:10.1038/nrgastro.2013.35 (12 March 2013).
Peterson, D.A., McNulty, N.P., Guruge, J.L. & Gordon, J.I. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2, 328–339 (2007).
Hapfelmeier, S. et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010).
Vaishnava, S. et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011).
Cash, H.L., Whitham, C.V., Behrendt, C.L. & Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006).
Brandl, K., Plitas, G., Schnabl, B., DeMatteo, R.P. & Pamer, E.G. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 204, 1891–1900 (2007).
Hugot, J.P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411, 599–603 (2001).
Ogura, Y. et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411, 603–606 (2001).
Wehkamp, J. et al. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut 53, 1658–1664 (2004).
Wehkamp, J. et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc. Natl. Acad. Sci. USA 102, 18129–18134 (2005).
Petnicki-Ocwieja, T. et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. USA 106, 15813–15818 (2009).
Jostins, L. et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Liu, C., Xu, Z., Gupta, D. & Dziarski, R. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J. Biol. Chem. 276, 34686–34694 (2001).
Saha, S. et al. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-gamma. Cell Host Microbe 8, 147–162 (2010).
Saha, S. et al. Peptidoglycan recognition proteins protect mice from experimental colitis by promoting normal gut flora and preventing induction of interferon-gamma. Cell Host Microbe 8, 147–162 (2010).
Bouskra, D. et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456, 507–510 (2008).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Mazmanian, S.K., Liu, C.H., Tzianabos, A.O. & Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005).
Mazmanian, S.K., Round, J.L. & Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Ochoa-Reparaz, J. et al. Central nervous system demyelinating disease protection by the human commensal Bacteroides fragilis depends on polysaccharide A expression. J. Immunol. 185, 4101–4108 (2010).
Round, J.L. & Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 107, 12204–12209 (2010).
Round, J.L. et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 (2011).
Shen, Y. et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12, 509–520 (2012).
Jeon, S.G. et al. Probiotic Bifidobacterium breve induces IL-10-producing Tr1 cells in the colon. PLoS Pathog. 8, e1002714 (2012).
Kirkland, D. et al. B cell-intrinsic MyD88 signaling prevents the lethal dissemination of commensal bacteria during colonic damage. Immunity 36, 228–238 (2012).
Manicassamy, S. & Pulendran, B. Modulation of adaptive immunity with Toll-like receptors. Semin. Immunol. 21, 185–193 (2009).
Fukata, M. et al. The myeloid differentiation factor 88 (MyD88) is required for CD4+ T cell effector function in a murine model of inflammatory bowel disease. J. Immunol. 180, 1886–1894 (2008).
Caramalho, I. et al. Regulatory T cells selectively express toll-like receptors and are activated by lipopolysaccharide. J. Exp. Med. 197, 403–411 (2003).
Komai-Koma, M., Jones, L., Ogg, G.S., Xu, D. & Liew, F.Y. TLR2 is expressed on activated T cells as a costimulatory receptor. Proc. Natl. Acad. Sci. USA 101, 3029–3034 (2004).
Liu, H., Komai-Koma, M., Xu, D. & Liew, F.Y. Toll-like receptor 2 signaling modulates the functions of CD4+ CD25+ regulatory T cells. Proc. Natl. Acad. Sci. USA 103, 7048–7053 (2006).
Sutmuller, R.P. et al. Toll-like receptor 2 controls expansion and function of regulatory T cells. J. Clin. Invest. 116, 485–494 (2006).
Clarke, T.B. et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 16, 228–231 (2010).
Hill, D.A. et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 18, 538–546 (2012).
Ichinohe, T. et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 108, 5354–5359 (2011).
Hooper, L.V. Do symbiotic bacteria subvert host immunity? Nat. Rev. Microbiol. 7, 367–374 (2009).
Acknowledgements
We thank A. Khosravi, S.W. McBride, G. Sharon, Y. Lee and M. Flajnik for comments on the manuscript. Supported by the Burroughs Wellcome Fund, Crohn's and Colitis Foundation and US National Institutes of Health (DK078938 and GM099535).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Chu, H., Mazmanian, S. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol 14, 668–675 (2013). https://doi.org/10.1038/ni.2635
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2635
This article is cited by
-
Pathophysiology of acute lung injury in patients with acute brain injury: the triple-hit hypothesis
Critical Care (2024)
-
Studies of the Impact of the Bifidobacterium Species on Inducible Nitric Oxide Synthase Expression and Nitric Oxide Production in Murine Macrophages of the BMDM Cell Line
Probiotics and Antimicrobial Proteins (2023)
-
Dysbiosis of the larval gut microbiota of Spodoptera frugiperda strains feeding on different host plants
Symbiosis (2023)
-
Interactions between Glossina pallidipes salivary gland hypertrophy virus and tsetse endosymbionts in wild tsetse populations
Parasites & Vectors (2022)
-
Timing matters: age-dependent impacts of the social environment and host selection on the avian gut microbiota
Microbiome (2022)