Abstract
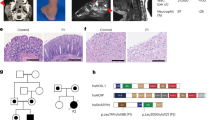
We report the clinical description and molecular dissection of a new fatal human inherited disorder characterized by chronic autoinflammation, invasive bacterial infections and muscular amylopectinosis. Patients from two kindreds carried biallelic loss-of-expression and loss-of-function mutations in HOIL1 (RBCK1), a component of the linear ubiquitination chain assembly complex (LUBAC). These mutations resulted in impairment of LUBAC stability. NF-κB activation in response to interleukin 1β (IL-1β) was compromised in the patients' fibroblasts. By contrast, the patients' mononuclear leukocytes, particularly monocytes, were hyper-responsive to IL-1β. The consequences of human HOIL-1 and LUBAC deficiencies for IL-1β responses thus differed between cell types, consistent with the unique association of autoinflammation and immunodeficiency in these patients. These data suggest that LUBAC regulates NF-κB–dependent IL-1β responses differently in different cell types.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Masters, S.L., Simon, A., Aksentijevich, I. & Kastner, D.L. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease. Annu. Rev. Immunol. 27, 621–668 (2009).
Aksentijevich, I. et al. An autoinflammatory disease with deficiency of the interleukin-1-receptor antagonist. N. Engl. J. Med. 360, 2426–2437 (2009).
McDermott, M.F. et al. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell 97, 133–144 (1999).
Ombrello, M.J. et al. Cold urticaria, immunodeficiency, and autoimmunity related to PLCG2 deletions. N. Engl. J. Med. 366, 330–338 (2012).
Puel, A., Picard, C., Ku, C.L., Smahi, A. & Casanova, J.L. Inherited disorders of NF-κB-mediated immunity in man. Curr. Opin. Immunol. 16, 34–41 (2004).
Doffinger, R. et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-κB signaling. Nat. Genet. 27, 277–285 (2001).
Courtois, G. et al. A hypermorphic IκBα mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J. Clin. Invest. 112, 1108–1115 (2003).
Casanova, J.L., Abel, L. & Quintana-Murci, L. Human TLRs and IL-1Rs in host defense: natural insights from evolutionary, epidemiological, and clinical genetics. Annu. Rev. Immunol. 29, 447–491 (2011).
von Bernuth, H. et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science 321, 691–696 (2008).
Picard, C. et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science 299, 2076–2079 (2003).
Picard, C. et al. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine 89, 403–425 (2010).
Casanova, J.L. & Abel, L. Inborn errors of immunity to infection: the rule rather than the exception. J. Exp. Med. 202, 197–201 (2005).
Casanova, J.L. & Abel, L. Primary immunodeficiencies: a field in its infancy. Science 317, 617–619 (2007).
Picard, C., Casanova, J.L. & Puel, A. Infectious diseases in patients with IRAK-4, MyD88, NEMO, or IκBα deficiency. Clin. Microbiol. Rev. 24, 490–497 (2011).
von Bernuth, H. et al. Septicemia without sepsis: inherited disorders of nuclear factor-κB-mediated inflammation. Clin. Infect. Dis. 41 (suppl. 7), S436–S439 (2005).
Tokunaga, F. et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132 (2009).
Iwai, K. & Tokunaga, F. Linear polyubiquitination: a new regulator of NF-κB activation. EMBO Rep. 10, 706–713 (2009).
Vissers, L.E., Veltman, J.A., van Kessel, A.G. & Brunner, H.G. Identification of disease genes by whole genome CGH arrays. Hum. Mol. Genet. 14, R215–R223 (2005).
Byun, M. et al. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J. Exp. Med. 207, 2307–2312 (2010).
Bolze, A. et al. Whole-exome-sequencing-based discovery of human FADD deficiency. Am. J. Hum. Genet. 87, 873–881 (2010).
Kirisako, T. et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 (2006).
Ikeda, F. et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471, 637–641 (2011).
Gerlach, B. et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 (2011).
Tokunaga, F. et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 (2011).
Tatematsu, K. et al. Transcriptional activity of RBCK1 protein (RBCC protein interacting with PKC 1): requirement of RING-finger and B-Box motifs and regulation by protein kinases. Biochem. Biophys. Res. Commun. 247, 392–396 (1998).
Tian, Y. et al. RBCK1 negatively regulates tumor necrosis factor- and interleukin-1-triggered NF-κB activation by targeting TAB2/3 for degradation. J. Biol. Chem. 282, 16776–16782 (2007).
Haas, T.L. et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36, 831–844 (2009).
Hostager, B.S., Kashiwada, M., Colgan, J.D. & Rothman, P.B. HOIL-1L interacting protein (HOIP) is essential for CD40 signaling. PLoS ONE 6, e23061 (2011).
Zak, D.E. et al. Systems analysis identifies an essential role for SHANK-associated RH domain-interacting protein (SHARPIN) in macrophage Toll-like receptor 2 (TLR2) responses. Proc. Natl. Acad. Sci. USA 108, 11536–11541 (2011).
Picard, C., Puel, A., Bustamante, J., Ku, C.L. & Casanova, J.L. Primary immunodeficiencies associated with pneumococcal disease. Curr. Opin. Allergy Clin. Immunol. 3, 451–459 (2003).
Seymour, R.E. et al. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 8, 416–421 (2007).
HogenEsch, H., Janke, S., Boggess, D. & Sundberg, J.P. Absence of Peyer's patches and abnormal lymphoid architecture in chronic proliferative dermatitis (cpdm/cpdm) mice. J. Immunol. 162, 3890–3896 (1999).
HogenEsch, H. et al. Increased expression of type 2 cytokines in chronic proliferative dermatitis (cpdm) mutant mice and resolution of inflammation following treatment with IL-12. Eur. J. Immunol. 31, 734–742 (2001).
Liang, Y., Seymour, R.E. & Sundberg, J.P. Inhibition of NF-kappaB signaling retards eosinophilic dermatitis in SHARPIN-deficient mice. J. Invest. Dermatol. 131, 141–149 (2011).
Moses, S.W. & Parvari, R. The variable presentations of glycogen storage disease type IV: a review of clinical, enzymatic and molecular studies. Curr. Mol. Med. 2, 177–188 (2002).
Bruno, C. et al. Clinical and genetic heterogeneity of branching enzyme deficiency (glycogenosis type IV). Neurology 63, 1053–1058 (2004).
Pellissier, J.F., de Barsy, T., Bille, J., Serratrice, G. & Toga, M. Polysaccharide (amylopectin-like) storage myopathy histochemical ultrastructural and biochemical studies. Acta Neuropathol. Suppl. 7, 292–296 (1981).
Ewert, R. et al. [Glycogenosis type IV as a rare cause of cardiomyopathy—report of a successful heart transplantation.] Z. Kardiol. 88, 850–856 (1999).
Vernia, S., Rubio, T., Heredia, M., Rodriguez de Cordoba, S. & Sanz, P. Increased endoplasmic reticulum stress and decreased proteasomal function in lafora disease models lacking the phosphatase laforin. PLoS ONE 4, e5907 (2009).
Lesca, G. et al. Novel mutations in EPM2A and NHLRC1 widen the spectrum of Lafora disease. Epilepsia 51, 1691–1698 (2010).
Monaghan, T.S. & Delanty, N. Lafora disease: epidemiology, pathophysiology and management. CNS Drugs 24, 549–561 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Smahi, A. et al. Genomic rearrangement in NEMO impairs NF-κB activation and is a cause of incontinentia pigmenti. The International Incontinentia Pigmenti (IP) Consortium. Nature 405, 466–472 (2000).
Chaussabel, D. et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity 29, 150–164 (2008).
Acknowledgements
We thank the children and their families for participating, and F. Iserin, V. Colomb, F. Rüemmele and V. Valayannopoulos for taking care of them. We particularly thank L. Abel, A. Durandy, P. Génin, B. Neven, M. Veron, J.W. Verbsky and R. Weil. H. Walczak (Imperial College London), K. Iwai (Osaka University) and A. Smahi (INSERM U781 Necker hospital, Paris Cité Sorbonne University) provided antibodies and cells. This work was partly funded by US National Center for Advancing Translational Sciences and National Center for Research Resources, US National Institutes of Health (NIH; 8UL1TR000043), St. Giles Foundation, Jeffrey Modell Foundation, Rockefeller University, INSERM, Paris Descartes University, US National Institute of Allergy and Infectious Diseases (R21AI085523; J.-L.C. and D.C.), NIH (5P01AI061093; J.-L.C.), NIH (R01AR050770; V.P.), Canceropole Ile de France (2007; A.I.), European Community Network of Excellence-Role of Ubiquitin and Ubiquitin-like Modifiers in Cellular Regulation (LSHC-CT-2005-018683; E.L. and A.I.), Thrasher Research Fund (C. Prando), Institut de Recherches Servier (E.L., F.A. and A.I.) and Manton Foundation (L.D.N.).
Author information
Authors and Affiliations
Contributions
B.B., E.L., S.G., A.A., L.I., G.T.-N. and M.C. performed experiments. C. Prando, A.A. and D.V. performed genetic analysis. F.B., M.D., E.M., D. Bonnet., P.Q., L.D.N. and C. Picard provided all the clinical data for the patients. D. Bogunovic., D.M., M.H., F.A. and H.W.V. provided reagents and suggestions. X.B. and C. Picard performed immunological explorations. C.R., F.F. and J.-C.F. performed histological analysis. E.I., Z.X., A.-M.C., V.P. and D.C. performed transcriptome analysis. A.I., J.-L.C. and C. Picard coordinated the study, and B.B., E.L., C. Prando, V.P., D.C., L.D.N., A.P., A.I., J.-L.C. and C. Picard wrote the manuscript. All authors discussed the results and commented on the manuscript. B.B., E.L. and C. Prando equally contributed as first authors. S.G., E.I., Z.X. and A.A. equally contributed as second authors. V.P., D.C., L.D.N. and A.P. equally contributed as second to last authors. A.I., J.-L.C. and C. Picard equally contributed as last authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–7 (PDF 3850 kb)
Rights and permissions
About this article
Cite this article
Boisson, B., Laplantine, E., Prando, C. et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol 13, 1178–1186 (2012). https://doi.org/10.1038/ni.2457
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2457
This article is cited by
-
RNF31 promotes tumorigenesis via inhibiting RIPK1 kinase-dependent apoptosis
Oncogene (2023)
-
HOIL1 regulates group 2 innate lymphoid cell numbers and type 2 inflammation in the small intestine
Mucosal Immunology (2022)
-
Heme as a differentiation-regulatory transcriptional cofactor
International Journal of Hematology (2022)
-
Polyglucosan body myopathy 1 may cause cognitive impairment: a case report from China
BMC Musculoskeletal Disorders (2021)
-
Dual roles for LUBAC signaling in thymic epithelial cell development and survival
Cell Death & Differentiation (2021)