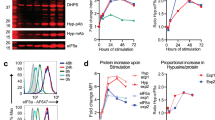
Abstract
Immunoglobulin secretion is modulated by competition between the use of a weak promoter-proximal poly(A) site and a nonconsensus splice site in the final secretory-specific exon of the heavy chain pre-mRNA. The RNA polymerase II transcription elongation factor ELL2, which is induced in plasma cells, enhanced both polyadenylation and exon skipping with the gene encoding the immunoglobulin heavy-chain complex (Igh) and reporter constructs. Lowering ELL2 expression by transfection of heterogenous ribonucleoprotein F (hnRNP F) or small interfering RNA resulted in lower abundance of secretory-specific forms of immunoglobulin heavy-chain mRNA. ELL2 and the polyadenylation factor CstF-64 tracked together with RNA polymerase II across the Igh μ- and γ-gene segments; the association of both factors was blocked by ELL2-specific small interfering RNA. Thus, loading of ELL2 and CstF-64 on RNA polymerase II was linked, caused enhanced use of the proximal poly(A) site and was necessary for processing of immunoglobulin heavy-chain mRNA.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Edwalds-Gilbert, G., Veraldi, K. & Milcarek, C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 25, 2547–2561 (1997).
Tian, B., Pan, Z. & Lee, J.Y. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 17, 156–165 (2007).
Shell, S.A., Martincic, K., Tran, J. & Milcarek, C. Increased phosphorylation of the carboxyl terminal domain of RNA polymerase II and loading of polyadenylation and co-transcriptional factors contribute to regulation of the Ig heavy chain mRNA in plasma cells. J. Immunol. 179, 7663–7673 (2007).
Kelly, D.E. & Perry, R.P. Transcriptional and post-transcriptional control of Ig mRNA production during B lymphocyte development. Nucleic Acids Res. 14, 5431–5441 (1986).
Thirman, M.J., Levitan, D.A., Kobayahi, H., Simon, M.C. & Rowley, J.D. Cloning of ELL, a gene that fuses to MLL in a t(11:19)(q23:p13.1) in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 91, 12110–12114 (1994).
Shilatifard, A., Lane, W.S., Jackson, K.W., Conaway, R.C. & Conaway, J.W. An RNA polymerase II elongation factor encoded by the human ELL gene. Science 271, 1873–1876 (1996).
Shilatifard, A. et al. ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc. Natl. Acad. Sci. USA 94, 3639–3643 (1997).
Miller, T., Williams, K., Johnstone, R.W. & Shilatifard, A. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J. Biol. Chem. 275, 32052–32056 (2000).
Sciammas, R. & Davis, M.M. Modular nature of Blimp-1 in the regulation of gene expression during B cell maturation. J. Immunol. 172, 5427–5440 (2004).
Shaffer, A.L. et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93 (2004).
Shaffer, A.L. et al. IRF4 addiction in multiple myeloma. Nature 454, 226–231 (2008).
Kornblihtt, A.R., De La Mata, M., Fededa, J.-P., Munoz, M.J. & Nogues, G. Multiple links between transcription and splicing. RNA 10, 1489–1498 (2004).
Rozenblatt-Rosen, O. et al. The tumor suppressor Cdc73 functionally associates with CPSF and CstF mRNA processing factors. Proc. Natl. Acad. Sci. USA 106, 755–760 (2009).
Veraldi, K.L. et al. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B-cells. Mol. Cell. Biol. 21, 1228–1238 (2001).
Alkan, S.A., Martincic, K. & Milcarek, C. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem. J. 393, 361–371 (2006).
Min, H., Chan, R. & Black, D. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 9, 2659–2671 (1995).
Milcarek, C., Hartman, M. & Croll, S. Changes in abundance of IgG 2a mRNA in the nucleus and cytoplasm of a murine B-lymphoma before and after fusion to a myeloma cell. Mol. Immunol. 33, 691–701 (1996).
Malhotra, J.D. & Kaufman, R.F. The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 18, 716–731 (2007).
Kobrin, B.J., Milcarek, C. & Morrison, S.L. Sequences near the 3′ secretion-specific polyadenylation site influence levels of secretion-specific and membrane-specific IgG2b mRNA in myeloma cells. Mol. Cell. Biol. 6, 1687–1697 (1986).
Takagaki, Y. & Manley, J.L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20, 1515–1525 (2000).
Qu, X. et al. The C-terminal domains of vertebrate CstF-64 and its yeast orthologue Rna15 form a new structure critical for mRNA 3′-end processing. J. Biol. Chem. 282, 2101–2115 (2007).
Calvo, C. & Manley, J.L. The transcriptional coactivator PC4/sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 24, 1009–1020 (2005).
Lewis, B.A., Sims, R.J. III, Lane, W.S. & Reinberg, D. Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol. Cell 18, 471–481 (2005).
Underhill, G.H., George, D., Bremer, E.G. & Kansas, G.S. Gene expression profiling reveals a highly specialized genetic program of plasma cells. Blood 101, 4013–4021 (2003).
Matis, S.A., Martincic, K. & Milcarek, C. B-lineage regulated polyadenylation occurs on weak poly(A) sites regardless of sequence composition at the cleavage and downstream regions. Nucleic Acids Res. 24, 4684–4692 (1996).
Seipelt, R.L. & Peterson, M.L. Alternative processing of IgA pre-mRNA responds like IgM to alterations in the efficiency of the competing splice and cleavage-polyadenylation reactions. Mol. Immunol. 32, 277–285 (1995).
Cramer, P., Pesce, C.G., Baralle, F.E. & Kornblihtt, A.R. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. USA 94, 11456–11460 (1997).
Cramer, P. et al. Coupling of transcription with alternative splicing: RNA polII promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol. Cell 4, 251–258 (1999).
de la Mata, M. et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12, 525–532 (2003).
Bruce, S.R., Dingle, R.W.C. & Peterson, M.L. B-cell and plasma-cell splicing differences: A potential role in regulated immunoglobulin RNA processing. RNA 9, 1264–1273 (2003).
Ma, J., Gunderson, S.I. & Phillips, C. Non-snRNP U1A levels decrease during mammalian B-cell differentiation and release the IgM secretory poly(A) site from repression. RNA 12, 122–132 (2006).
Takagaki, Y. & Manley, J. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell 2, 761–771 (1998).
Peterson, M.L. & Perry, R.P. The regulated production of μ-m and μ-s mRNA is dependent on the relative efficiencies of μ-s poly(A) site usage and the Cmu4 to M1 splice. Mol. Cell. Biol. 9, 726–738 (1989).
Auboeuf, D. et al. A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol. Cell. Biol. 25, 5307–5316 (2005).
Shi, Y. et al. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33, 365–376 (2009).
Rigo, F. & Martinson, H.G. Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol. Cell. Biol. 28, 849–862 (2008).
Venkataraman, K., Brown, K.M. & Gilmartin, G.M. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 19, 1315–1327 (2005).
Glover-Cutter, K., Kim, S., Espinosa, J. & Bentley, D.L. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 15, 71–78 (2008).
Gendra, E., Colgan, D.F., Meany, B. & Konarska, M.M. A sequence motif in the simian virus 40 (SV40) early core promoter affects alternative splicing of transcribed mRNA. J. Biol. Chem. 282, 11648–11657 (2007).
Tusher, V.G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121 (2001).
Edwalds-Gilbert, G. & Milcarek, C. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol. Cell. Biol. 15, 6420–6429 (1995).
Shell, S.A., Hesse, C., Morris, S.M., Jr & Milcarek, C. Elevated levels of the 64-kDa ceavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J. Biol. Chem. 280, 39950–39961 (2005).
Milcarek, C. & Hall, B. Cell-specific expression of secreted versus membrane forms of immunoglobulin γ2b mRNA involves selective use of alternate polyadenylation sites. Mol. Cell. Biol. 5, 2514–2520 (1985).
Acknowledgements
We thank A. Shilatifard (Stowers Institute for Medical Research) for monoclonal antibody to ELL2; A. Kornblihtt (University of Buenos Aires) for splicing reporter plasmids; C. Webb (University of Oklahoma) for Igh promoters; J. Fink for assistance in constructing COOH ELL2; S. Eghtesad for cloning ELL2-specific shRNA; J. Heakins for mouse spleen cell chromatin; and B. Junker and Y. Lin for help with statistical analyses. Supported by the US National Institutes of Health (CA86433 to C.M. and AI079047 to L.B.) and the National Science Foundation (MCB-8842725 to C.M.).
Author information
Authors and Affiliations
Contributions
K.M. cloned ELL2 segments for Figure 1b and did immunoblot analyses for Figure 2, transfection for Figures 5a, 6 and 7 and in vitro studies for Supplementary Figures 1 and 2 and prepared all figures; S.A.A. did the microarray study; A.C. did the quantitative RT-PCR for Table 1; L.B. provided the splenic B cells for Figures 2c, 3a and 5b; C.M. did the RNA hybridization blot for Figure 2a, the ChIP experiments for Figures 3 and 4 and the quantitative RT-PCR for Figure 5b, and conceived the studies, provided critical guidance and wrote the manuscript with assistance from K.M. and L.B; and all authors reviewed and approved the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1–2 and Supplementary Methods (PDF 509 kb)
Rights and permissions
About this article
Cite this article
Martincic, K., Alkan, S., Cheatle, A. et al. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol 10, 1102–1109 (2009). https://doi.org/10.1038/ni.1786
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.1786
This article is cited by
-
Functional dissection of inherited non-coding variation influencing multiple myeloma risk
Nature Communications (2022)
-
Genetic predisposition for multiple myeloma
Leukemia (2020)
-
Multivariate discovery and replication of five novel loci associated with Immunoglobulin G N-glycosylation
Nature Communications (2017)
-
Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response
Nature Immunology (2016)
-
Multifunctional role of the transcription factor Blimp-1 in coordinating plasma cell differentiation
Nature Immunology (2016)