Abstract
Cancers are characterized by non-random chromosome copy number alterations that presumably contain oncogenes and tumor-suppressor genes (TSGs). The affected loci are often large, making it difficult to pinpoint which genes are driving the cancer. Here we report a cross-species in vivo screen of 84 candidate oncogenes and 39 candidate TSGs, located within 28 recurrent chromosomal alterations in ependymoma. Through a series of mouse models, we validate eight new ependymoma oncogenes and ten new ependymoma TSGs that converge on a small number of cell functions, including vesicle trafficking, DNA modification and cholesterol biosynthesis, identifying these as potential new therapeutic targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Schwab, M. et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 305, 245–248 (1983).
Nobori, T. et al. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 368, 753–756 (1994).
Zender, L. et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell 135, 852–864 (2008).
Johnson, R.A. et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature 466, 632–636 (2010).
Parker, M. et al. C11orf95-RELA fusions drive oncogenic NF-κB signaling in ependymoma. Nature 506, 451–455 (2014).
Atkinson, J.M. et al. An integrated in vitro and in vivo high-throughput screen identifies treatment leads for ependymoma. Cancer Cell 20, 384–399 (2011).
Mack, S.C. et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 506, 445–450 (2014).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Chautard, E., Ouédraogo, Z., Biau, J. & Verrelle, P. Role of Akt in human malignant glioma: from oncogenesis to tumor aggressiveness. J. Neurooncol. 117, 205–215 (2014).
Hu, R., Wang, E., Peng, G., Dai, H. & Lin, S.-Y. Zinc finger protein 668 interacts with Tip60 to promote H2AX acetylation after DNA damage. Cell Cycle 12, 2033–2041 (2013).
Kadoch, C. et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601 (2013).
Müller, M., Pym, E.C., Tong, A. & Davis, G.W. Rab3-GAP controls the progression of synaptic homeostasis at a late stage of vesicle release. Neuron 69, 749–762 (2011).
Tian, M., Xu, C.S., Montpetit, R. & Kramer, R.H. Rab3A mediates vesicle delivery at photoreceptor ribbon synapses. J. Neurosci. 32, 6931–6936 (2012).
Lu, W. et al. Peroxiredoxin 2 is upregulated in colorectal cancer and contributes to colorectal cancer cells' survival by protecting cells from oxidative stress. Mol. Cell. Biochem. 387, 261–270 (2014).
Donson, A.M. et al. Immune gene and cell enrichment is associated with a good prognosis in ependymoma. J. Immunol. 183, 7428–7440 (2009).
Lin, W. et al. Mammalian DNA2 helicase/nuclease cleaves Gquadruplex DNA and is required for telomere integrity. EMBO J. 32, 1425–1439 (2013).
Anazawa, Y., Arakawa, H., Nakagawa, H. & Nakamura, Y. Identification of STAG1 as a key mediator of a p53-dependent apoptotic pathway. Oncogene 23, 7621–7627 (2004).
Hsu, C.-H. et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep. 2, 568–579 (2012).
Poage, G.M. et al. Identification of an epigenetic profile classifier that is associated with survival in head and neck cancer. Cancer Res. 72, 2728–2737 (2012).
Zhou, X. et al. Unc-51–like kinase 1/2–mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc. Natl. Acad. Sci. USA 104, 5842–5847 (2007).
Yang, H., Sasaki, T., Minoshima, S. & Shimizu, N. Identification of three novel proteins (SGSM1, 2, 3) which modulate small G protein (RAP and RAB)-mediated signaling pathway. Genomics 90, 249–260 (2007).
Wassmer, T. et al. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev. Cell 17, 110–122 (2009).
Moughamian, A.J., Osborn, G.E., Lazarus, J.E., Maday, S. & Holzbaur, E.L.F. Ordered recruitment of dynactin to the microtubule plus-end is required for efficient initiation of retrograde axonal transport. J. Neurosci. 33, 13190–13203 (2013).
Eggers, C.T., Schafer, J.C., Goldenring, J.R. & Taylor, S.S. D-AKAP2 interacts with Rab4 and Rab11 through its RGS domains and regulates transferrin receptor recycling. J. Biol. Chem. 284, 32869–32880 (2009).
Wojtal, K.A., de Vries, E., Hoekstra, D. & van IJzendoorn, S.C.D. Efficient trafficking of MDR1/P-glycoprotein to apical canalicular plasma membranes in HepG2 cells requires PKA-RIIα anchoring and glucosylceramide. Mol. Biol. Cell 17, 3638–3650 (2006).
Remeseiro, S. et al. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 31, 2076–2089 (2012).
Tsutsui, T. et al. Mediator complex recruits epigenetic regulators via its two cyclin-dependent kinase subunits to repress transcription of immune response genes. J. Biol. Chem. 288, 20955–20965 (2013).
Goldenring, J.R. A central role for vesicle trafficking in epithelial neoplasia: intracellular highways to carcinogenesis. Nat. Rev. Cancer 13, 813–820 (2013).
Mosesson, Y., Mills, G.B. & Yarden, Y. Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer 8, 835–850 (2008).
Sie, M. et al. Growth-factor-driven rescue to receptor tyrosine kinase (RTK) inhibitors through Akt and Erk phosphorylation in pediatric low grade astrocytoma and ependymoma. PLoS ONE 10, e0122555 (2015).
Sikkema, A.H. et al. Kinome profiling in pediatric brain tumors as a new approach for target discovery. Cancer Res. 69, 5987–5995 (2009).
Gaist, D. et al. Use of statins and risk of glioma: a nationwide case-control study in Denmark. Br. J. Cancer 108, 715–720 (2013).
Gaist, D., Hallas, J., Friis, S., Hansen, S. & Sørensen, H.T. Statin use and survival following glioblastoma multiforme. Cancer Epidemiol. 38, 722–727 (2014).
Nam, K.T. et al. Loss of Rab25 promotes the development of intestinal neoplasia in mice and is associated with human colorectal adenocarcinomas. J. Clin. Invest. 120, 840–849 (2010).
Morrison, H.A. et al. Regulation of early endosomal entry by the Drosophila tumor suppressors Rabenosyn and Vps45. Mol. Biol. Cell 19, 4167–4176 (2008).
Tabori, U. et al. Human telomere reverse transcriptase expression predicts progression and survival in pediatric intracranial ependymoma. J. Clin. Oncol. 24, 1522–1528 (2006).
Zani, V.J. et al. Molecular cloning of complex chromosomal translocation t(8;14;12)(q24.1;q32.3;q24.1) in a Burkitt lymphoma cell line defines a new gene (BCL7A) with homology to caldesmon. Blood 87, 3124–3134 (1996).
Sigismund, S. et al. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev. Cell 15, 209–219 (2008).
Acknowledgements
We are grateful to F.B. Gertler (Massachusetts Institute of Technology) and S. Gupton (University of North Carolina) for the generous gift of the VAMP7-phlorin construct and the staffs of the Hartwell Center for Bioinformatics and Biotechnology, the Small Animal Imaging Center, the Animal Resources Center, the Cell and Tissue Imaging Center, and the Flow Cytometry and Cell Sorting Shared Resource at St. Jude Children's Research Hospital for technical assistance. This work was supported by grants from the US National Institutes of Health (R01CA129541, P01CA96832 and P30CA021765, R.J.G.), by the Collaborative Ependymoma Research Network (CERN) and by the American Lebanese Syrian Associated Charities (ALSAC).
Author information
Authors and Affiliations
Contributions
K.M.M. and D.S.C. conducted the great majority of the experiments and contributed to their design. E.W., N.B., J.D., C.E., B.N., R.T., M.C. and T.A.K. conducted mouse experimental work. G.N., S.O., Y.-D.W. and D.F. conducted microarray and deep sequencing studies. K.W. assisted with human genomic analyses. K.G. and D.W.E. conducted all neuropathological reviews. A.O.T. conducted all statistical analyses. R.J.G. conceived, designed and oversaw the study. All authors contributed to writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
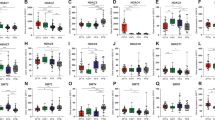
Supplementary Figure 1 Relative gain and loss of loci harboring ependymoma candidate oncogenes and TSGs in human cancers.
Heat maps report the relative gain and loss of loci harboring candidate ependymoma oncogenes and TSGs, respectively, across 3,116 samples of 9 different human tumors reported on the COSMIC database.
Supplementary Figure 2 Relative gain and loss of validated oncogenes and TSGs and non-validated candidates in human cancer.
Relative gain and loss of validated oncogenes and TSGs and non-validated candidates across 3,116 samples of 9 different human tumors reported on the COSMIC database (boxes extend from the 25th to 75th percentiles and contain a horizontal line marking the median value; whiskers extend to the maximum and minimum values).
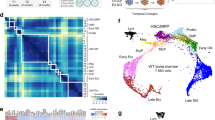
Supplementary Figure 3 AGDEX analysis of human and mouse ependymomas.
AGDEX analysis comparing the transcriptomic similarities of human cerebral ependymoma positive or negative for C11orf95-RELA fusion with our three new mouse models of cerebral ependymoma driven by BCL7C, RAB3A or ZNF668.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3 and Supplementary Note. (PDF 318 kb)
Supplementary Table 1
Candidate ependymoma oncogenes detected by integrated gene expression and copy number analysis of human ependymoma. (XLS 41 kb)
Supplementary Table 2
Candidate ependymoma tumor-suppressor genes detected by integrated gene expression and copy number analysis of human ependymoma. (XLS 54 kb)
Supplementary Table 3
Diploid control genes that are not amplified or overexpressed in human ependymoma. (XLS 33 kb)
Supplementary Table 4
Details of individual mice harboring pools 1–11 of NSCs carrying candidate oncogenes. (XLS 67 kb)
Supplementary Table 5
Details of individual mice harboring NSCs carrying diploid control genes. (XLS 36 kb)
Supplementary Table 6
Details of individual mice harboring NSCs carrying singlecandidate oncogenes. (XLS 96 kb)
Supplementary Table 7
The top most positively and negatively enriched gene sets common to mouse ependymomas driven by RAB3A, BCL7C and ZNF668 relative to cNSCs, detected by GSEA. (XLS 95 kb)
Supplementary Table 8
Details of individual mice harboring pools 1–4 of EPHB2-transduced cNSCs carrying shRNAs targeting candidate TSGs. (XLS 43 kb)
Supplementary Table 9
Validated ependymoma oncogenes and tumor-suppressor genes identified in in vivo screens. (XLS 33 kb)
Rights and permissions
About this article
Cite this article
Mohankumar, K., Currle, D., White, E. et al. An in vivo screen identifies ependymoma oncogenes and tumor-suppressor genes. Nat Genet 47, 878–887 (2015). https://doi.org/10.1038/ng.3323
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3323
This article is cited by
-
Identification of cell subpopulations associated with disease phenotypes from scRNA-seq data using PACSI
BMC Biology (2023)
-
Superenhancers as master gene regulators and novel therapeutic targets in brain tumors
Experimental & Molecular Medicine (2023)
-
Genome-wide CRISPR knockout screening identified G protein pathway suppressor 2 as a novel tumor suppressor for uveal melanoma metastasis
Journal of Cancer Research and Clinical Oncology (2023)
-
The biology of ependymomas and emerging novel therapies
Nature Reviews Cancer (2022)
-
Epigenetic Alterations of Repeated Relapses in Patient-matched Childhood Ependymomas
Nature Communications (2022)