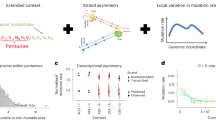
Abstract
Mutations create variation in the population, fuel evolution and cause genetic diseases. Current knowledge about de novo mutations is incomplete and mostly indirect1,2,3,4,5,6,7,8,9,10. Here we analyze 11,020 de novo mutations from the whole genomes of 250 families. We show that de novo mutations in the offspring of older fathers are not only more numerous11,12,13 but also occur more frequently in early-replicating, genic regions. Functional regions exhibit higher mutation rates due to CpG dinucleotides and show signatures of transcription-coupled repair, whereas mutation clusters with a unique signature point to a new mutational mechanism. Mutation and recombination rates independently associate with nucleotide diversity, and regional variation in human-chimpanzee divergence is only partly explained by heterogeneity in mutation rate. Finally, we provide a genome-wide mutation rate map for medical and population genetics applications. Our results provide new insights and refine long-standing hypotheses about human mutagenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sawyer, S.A. & Hartl, D.L. Population genetics of polymorphism and divergence. Genetics 132, 1161–1176 (1992).
Felsenstein, J. & Churchill, G.A. A hidden Markov model approach to variation among sites in rate of evolution. Mol. Biol. Evol. 13, 93–104 (1996).
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005).
Cooper, G.M. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15, 901–913 (2005).
Veltman, J.A. & Brunner, H.G. De novo mutations in human genetic disease. Nat. Rev. Genet. 13, 565–575 (2012).
Friedberg, E.C., Walker, G.C. & Siede, W. DNA Repair and Mutagenesis (ASM Press, 1995).
Kondrashov, A.S. Direct estimates of human per nucleotide mutation rates at 20 loci causing mendelian diseases. Hum. Mutat. 21, 12–27 (2003).
Lynch, M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA 107, 961–968 (2010).
Hodgkinson, A. & Eyre-Walker, A. Variation in the mutation rate across mammalian genomes. Nat. Rev. Genet. 12, 756–766 (2011).
Schaibley, V.M. et al. The influence of genomic context on mutation patterns in the human genome inferred from rare variants. Genome Res. 23, 1974–1984 (2013).
Michaelson, J.J. et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151, 1431–1442 (2012).
Kong, A. et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature 488, 471–475 (2012).
Genomes of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 46, 818–825 (2014).
Jenkins, T.G., Aston, K.I., Pflueger, C., Cairns, B.R. & Carrell, D.T. Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet. 10, e1004458 (2014).
Koren, A. DNA replication timing: coordinating genome stability with genome regulation on the X chromosome and beyond. Bioessays 36, 997–1004 (2014).
Arndt, P.F., Petrov, D.A. & Hwa, T. Distinct changes of genomic biases in nucleotide substitution at the time of Mammalian radiation. Mol. Biol. Evol. 20, 1887–1896 (2003).
Schmidt, S. et al. Hypermutable non-synonymous sites are under stronger negative selection. PLoS Genet. 4, e1000281 (2008).
Subramanian, S. & Kumar, S. Neutral substitutions occur at a faster rate in exons than in noncoding DNA in primate genomes. Genome Res. 13, 838–844 (2003).
Polak, P. et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat. Biotechnol. 32, 71–75 (2014).
Pleasance, E.D. et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature 463, 184–190 (2010).
Campbell, C.D. et al. Estimating the human mutation rate using autozygosity in a founder population. Nat. Genet. 44, 1277–1281 (2012).
Roberts, S.A. et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell 46, 424–435 (2012).
Nik-Zainal, S. et al. Mutational processes molding the genomes of 21 breast cancers. Cell 149, 979–993 (2012).
Chan, K., Resnick, M.A. & Gordenin, D.A. The choice of nucleotide inserted opposite abasic sites formed within chromosomal DNA reveals the polymerase activities participating in translesion DNA synthesis. DNA Repair (Amst.) 12, 878–889 (2013).
Arndt, P.F., Hwa, T. & Petrov, D.A. Substantial regional variation in substitution rates in the human genome: importance of GC content, gene density, and telomere-specific effects. J. Mol. Evol. 60, 748–763 (2005).
Hellmann, I., Ebersberger, I., Ptak, S.E., Pääbo, S. & Przeworski, M. A neutral explanation for the correlation of diversity with recombination rates in humans. Am. J. Hum. Genet. 72, 1527–1535 (2003).
Begun, D.J. & Aquadro, C.F. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature 356, 519–520 (1992).
Lercher, M.J. & Hurst, L.D. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 18, 337–340 (2002).
Kong, A. et al. A high-resolution recombination map of the human genome. Nat. Genet. 31, 241–247 (2002).
Duret, L. & Arndt, P.F. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 4, e1000071 (2008).
Kong, A. et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467, 1099–1103 (2010).
McVicker, G., Gordon, D., Davis, C. & Green, P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 5, e1000471 (2009).
Asthana, S., Roytberg, M., Stamatoyannopoulos, J. & Sunyaev, S. Analysis of sequence conservation at nucleotide resolution. PLOS Comput. Biol. 3, e254 (2007).
Gratten, J., Visscher, P.M., Mowry, B.J. & Wray, N.R. Interpreting the role of de novo protein-coding mutations in neuropsychiatric disease. Nat. Genet. 45, 234–238 (2013).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Koren, A. et al. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am. J. Hum. Genet. 91, 1033–1040 (2012).
Lappalainen, T. et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501, 506–511 (2013).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Ryba, T. et al. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res. 20, 761–770 (2010).
Kent, W.J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Paten, B., Herrero, J., Beal, K., Fitzgerald, S. & Birney, E. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 18, 1814–1828 (2008).
Flicek, P. et al. Ensembl 2013. Nucleic Acids Res. 41, D48–D55 (2013).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2014).
Blanchette, M. et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14, 708–715 (2004).
Murphy, W.J. et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294, 2348–2351 (2001).
Acknowledgements
We thank D. Gordenin for very helpful comments. The Genome of the Netherlands (GoNL) Project is funded by the Biobanking and Biomolecular Research Infrastructure (BBMRI-NL), which is financed by the Netherlands Organization for Scientific Research (NWO project 184.021.007). S.R.S., P.P.P. and S.C. are funded by US National Institutes of Health grants 1 R01 MH101244 and 1 R01 GM078598.
Author information
Authors and Affiliations
Consortia
Contributions
S.R.S. and P.I.W.d.B. planned and directed the research. L.C.F. called and filtered the mutations. W.P.K. and I.R. validated candidate mutations. L.C.F. designed and executed the simulations. A.K., L.C.F. and A.M. performed replication timing analyses. P.P.P., L.C.F. and A.M. analyzed factors influencing regional mutation rates and spectra. L.C.F. and P.P.P. analyzed mutation clusters. P.P.P. and P.F.A. computed the comparative genomics model and compared it against observed mutation rates. S.C. and P.P.P. created the mutation rate map. L.C.F., P.P.P., A.K., P.I.W.d.B. and S.R.S. wrote the manuscript. A.M., S.C., C.M.v.D., M.S., C.W., G.v.O., P.E.S., D.I.B., K.Y., V.G., P.F.A. and W.P.K. provided critical feedback on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of members and affiliations appears in the Supplementary Note.
Integrated supplementary information
Supplementary Figure 1 Association of paternal age of de novo mutation location with epigenetic variables.
Using a linear regression model, we tested the association of seven epigenetic variables (DNA replication timing, expression levels, recombination rates, and the H3K27ac, H3K4me1 and H3K4me3 histone modifications), while correcting for GC content, CpG status and sequence coverage. Here we plot the significance of the associations we found with the different epigenetic variables along with the significance threshold level after Bonferroni correction for the six tests we performed (gray dashed line).
Supplementary Figure 2 Separation of replication timing profiles between the offspring of younger and older fathers.
We separated our de novo mutation data in two groups based on paternal age. To select a threshold that maximizes the difference in the groups, we considered every integer age in our study as a possible threshold and applied a Kolmogorov-Smirnoff test to compare the distribution of replication timing of de novo mutations between the groups. This plot shows the P values obtained for each of the 27 tests as well as the significance threshold after Bonferroni correction (gray dashed line).
Supplementary Figure 3 Paternal age effect on de novo mutation replication timing measured in six cell types.
The distribution of replication timing around de novo mutations in the offspring of younger fathers (orange curves; <28 years old), older fathers (blue curves; ≥28 years old) and simulations (gray areas; 200 simulation sets of 11,020 mutations) in 3 cell types from 4 cell lines41: embryonic stem cells (BG01 ESC, H7 ESC), induced pluripotent stem cells (iPS4) and neural precursor cells (BG01 NPC).
Supplementary Figure 4 Paternal and maternal age effect estimates on replication timing based on de novo mutations with known parental origin.
All effect estimates were computed using a linear regression model. The red line represents the effect estimate from the 630 maternal mutations. The black curve shows the effect estimate for 10,000 samplings of 630 paternal mutations out of the 1,991 available. The effect estimate is significantly larger for paternal mutations (P = 0.0019) when considering the same number of mutations.
Supplementary Figure 5 Power to detect non-CpG mutation depletion in regulatory regions marked by DNase I–hypersensitive sites (DHSs).
The graph shows the power to detect non-CpG mutation depletion in regulatory regions marked by DNase I–hypersensitive sites (DHSs) using the 9,048 non-CpG observed de novo mutations in our data and 177,347 uniformly simulated mutations. The y axis illustrates the power for detecting an effect at the 0.05 significance level using a χ2 test for different effect sizes (here expressed as the relative depletion of mutations in DHSs when compared to other regions).
Supplementary Figure 6 Comparison of nucleotide context–specific mutation rates based on comparative genomics and observed de novo mutations.
Each point represents one of the 96 substitutions in a specific trinucleotide context. The black line shows the best fit (r2 = 0.993). The rate of transition mutations is 2.15 times greater than the transversion rate (Ti/Tv ratio). The highest mutation rates are observed for cytosine bases in a CpG context.
Supplementary Figure 7 Mutational spectrum in transcribed regions.
The proportion of de novo mutations of each substitution type in transcribed regions classified based on their corresponding strand (transcribed or non-transcribed). There is a strong asymmetry of mutations between the two strands, with significantly elevated A>G substitutions on the transcribed strand, consistent with the action of transcription-coupled nucleotide excision repair.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Tables 1–4 and Supplementary Note. (PDF 1185 kb)
Rights and permissions
About this article
Cite this article
Francioli, L., Polak, P., Koren, A. et al. Genome-wide patterns and properties of de novo mutations in humans. Nat Genet 47, 822–826 (2015). https://doi.org/10.1038/ng.3292
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3292
This article is cited by
-
Meta-analysis of 46,000 germline de novo mutations linked to human inherited disease
Human Genomics (2024)
-
Emergence and influence of sequence bias in evolutionarily malleable, mammalian tandem arrays
BMC Biology (2023)
-
Male infertility
Nature Reviews Disease Primers (2023)
-
Evolution of the germline mutation rate across vertebrates
Nature (2023)
-
Maximizing the value of twin studies in health and behaviour
Nature Human Behaviour (2023)