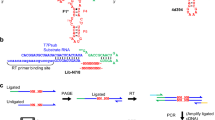
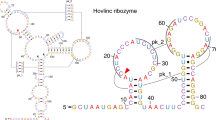
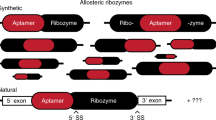
Abstract
Enzymes made of RNA catalyze reactions that are essential for protein synthesis and RNA processing. However, such natural ribozymes are exceedingly rare, as evidenced by the fact that the discovery rate for new classes has dropped to one per decade from about one per year during the 1980s. Indeed, only 11 distinct ribozyme classes have been experimentally validated to date. Recently, we recognized that self-cleaving ribozymes frequently associate with certain types of genes from bacteria. Herein we exploited this association to identify divergent architectures for two previously known ribozyme classes and to discover additional noncoding RNA motifs that are self-cleaving RNA candidates. We identified three new self-cleaving classes, which we named twister sister, pistol and hatchet, from this collection, suggesting that even more ribozymes remain hidden in modern cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Benner, S.A., Ellington, A.D. & Tauer, A. Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. USA 86, 7054–7058 (1989).
Nissen, P., Hansen, J., Ban, N., Moore, P.B. & Steitz, T.A. The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000).
Lambowitz, A.M., Caprara, M.G., Zimmerly, S. & Perlman, P.S. Group I and group II ribozymes as RNPs: clues to the past and guides to the future. in The RNA World edn. 2, vol. 37 (eds. Gesteland, R.F., Cech, T.R. & Atkins, J.F.) 451–483 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 1999).
Ellis, J.C. & Brown, J.W. The RNase P family. RNA Biol. 6, 362–369 (2009).
Ferré-D'Amaré, A.R. & Scott, W.G. Small self-cleaving ribozymes. Cold Spring Harb. Perspect. Biol. 2, a003574 (2010).
Perreault, J. et al. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput. Biol. 7, e1002031 (2011).
Webb, C.-H.T. & Lupták, A. HDV-like self-cleaving ribozymes. RNA Biol. 8, 719–727 (2011).
Roth, A. et al. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 10, 56–60 (2014).
Hutchins, C.J., Rathjen, P.D., Forster, A.C. & Symons, R.H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 14, 3627–3640 (1986).
Saville, B.J. & Collins, R.A. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell 61, 685–696 (1990).
Winkler, W.C., Nahvi, A., Roth, A., Collins, J.A. & Breaker, R.R. Control of gene expression by a natural metabolite–responsive ribozyme. Nature 428, 281–286 (2004).
Salehi-Ashtiani, K., Lupták, A., Litovchick, A. & Szostak, J.W. A genome-wide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313, 1788–1792 (2006).
Marchler-Bauer, A. et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 (2011).
Johnson, L.S., Eddy, S.R. & Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11, 431 (2010).
Yao, Z., Weinberg, Z. & Ruzzo, W.L. CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics 22, 445–452 (2006).
Weinberg, Z. et al. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 11, R31 (2010).
Pace, N.R., Thomas, B.C. & Woese, C.R. Probing RNA structure, function, and history by comparative analysis. Cold Spring Harb. Perspect. Biol. 37, 113–141 (1999).
Michel, F. & Westhof, E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 216, 585–610 (1990).
Weinberg, Z., Perreault, J., Meyer, M.M. & Breaker, R.R. Exceptional structured noncoding RNAs revealed by bacterial metagenome analysis. Nature 462, 656–659 (2009).
Westhof, E. The amazing world of bacterial structured RNAs. Genome Biol. 11, 108 (2010).
Sánchez-Luque, F.J., Lopez, M.C., Macias, F., Alonso, C. & Thomas, M.C. Identification of an hepatitis delta virus-like ribozyme at the mRNA 5′-end of the L1Tc retrotransposon from Trypanosoma cruzi. Nucleic Acids Res. 39, 8065–8077 (2011).
Breaker, R.R. et al. A common speed limit for RNA-cleaving ribozymes and deoxyribozymes. RNA 9, 949–957 (2003).
Liu, Y., Wilson, T.J., McPhee, S.A. & Lilley, D.M.J. Crystal structure and mechanistic investigation of the twister ribozyme. Nat. Chem. Biol. 10, 739–744 (2014).
Eiler, D., Wang, J. & Steitz, T.A. Structural basis for the fast self-cleavage reaction catalyzed by the twister ribozyme. Proc. Natl. Acad. Sci. USA 111, 13028–13033 (2014).
Ren, A. et al. In-line alignment and Mg2+ coordination at the cleavage site of the env22 twister ribozyme. Nat. Commun. 5, 5534–5543 (2014).
Tang, J. & Breaker, R.R. Structural diversity of self-cleaving ribozymes. Proc. Natl. Acad. Sci. USA 97, 5784–5789 (2000).
Salehi-Ashtiani, K. & Szostak, J.W. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414, 82–84 (2001).
Pruitt, K.D., Tatusova, T. & Maglott, D.R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65 (2007).
Markowitz, V.M. et al. IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res. 40, D123–D129 (2012).
Human Microbiome Project Consortium. A framework for human microbiome research. Nature 486, 215–221 (2012).
Meyer, F. et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 (2008).
Sun, S. et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res. 39, D546–D551 (2011).
Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J. & Wheeler, D.L. GenBank. Nucleic Acids Res. 36, D25–D30 (2008).
Noguchi, H., Park, J. & Takagi, T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 34, 5623–5630 (2006).
Zhu, W., Lomsadze, A. & Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38, e132 (2010).
Baker, J.L. et al. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 335, 233–235 (2012).
Nawrocki, E.P. & Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013).
Burge, S.W. et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 41, D226–D232 (2013).
Lowe, T.M. & Eddy, S.R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Bland, C. et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8, 209 (2007).
Huang, Y., Gilna, P. & Li, W. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25, 1338–1340 (2009).
Gardner, P.P., Barquist, L., Bateman, A., Nawrocki, E.P. & Weinberg, Z. RNIE: genome-wide prediction of bacterial intrinsic terminators. Nucleic Acids Res. 39, 5845–5852 (2011).
Weinberg, Z. & Breaker, R.R. R2R—software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12, 3 (2011).
Ruminski, D.J., Webb, C.H., Riccitelli, N.J. & Luptak, A. Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J. Biol. Chem. 286, 41286–41295 (2011).
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010).
Acknowledgements
We thank A. Roth and other members of the Breaker laboratory for helpful discussions, and N. Carriero and R. Bjornson for assistance with the Yale Life Sciences High Performance Computing Center funded by the US National Institutes of Health (NIH; RR19895-02). P.B.K. was supported by an NIH Genetics Training Grant (5T32GM007499). C.E.L. was supported by the Deutsche Forschungsgemeinschaft (LU1889/1-1). This work was also supported by an NIH grant (GM022778) to R.R.B. and by the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
Z.W. conducted all bioinformatics searches and created the RNA consensus models. Validation studies were conducted for twister sister by P.B.K. and T.H.C., for pistol by K.A.H., for hatchet by S.L., for hammerhead by P.B.K. and for HDV by P.B.K. and S.L. P.B.K., T.H.C. and C.E.L. screened additional noncleaving RNAs. R.R.B. worked with all authors to plan experiments, interpret data and write the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–5 and Supplementary Figures 1–13 (PDF 2888 kb)
Supplementary Data Set 1
Printable alignments, genomic contexts and taxonomy of self-cleaving ribozyme candidates (PDF 6312 kb)
Supplementary Data Set 2
Machine-readable alignments of self-cleaving ribozyme candidates (ZIP 125 kb)
Rights and permissions
About this article
Cite this article
Weinberg, Z., Kim, P., Chen, T. et al. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol 11, 606–610 (2015). https://doi.org/10.1038/nchembio.1846
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1846
This article is cited by
-
Deep generative design of RNA family sequences
Nature Methods (2024)
-
Theoretical Studies of the Self Cleavage Pistol Ribozyme Mechanism
Topics in Catalysis (2022)
-
Nutzung von RNA-seq zur Detektion aktiver Ribozyme in Zellextrakten
BIOspektrum (2022)
-
Identification of 11 candidate structured noncoding RNA motifs in humans by comparative genomics
BMC Genomics (2021)
-
Comparative genomics identifies thousands of candidate structured RNAs in human microbiomes
Genome Biology (2021)