Abstract
Leukaemogenesis requires enhanced self-renewal, which is induced by oncogenes. The underlying molecular mechanisms remain incompletely understood. Here, we identified C/D box snoRNAs and rRNA 2′-O-methylation as critical determinants of leukaemic stem cell activity. Leukaemogenesis by AML1-ETO required expression of the groucho-related amino-terminal enhancer of split (AES). AES functioned by inducing snoRNA/RNP formation via interaction with the RNA helicase DDX21. Similarly, global loss of C/D box snoRNAs with concomitant loss of rRNA 2′-O-methylation resulted in decreased leukaemia self-renewal potential. Genomic deletion of either C/D box snoRNA SNORD14D or SNORD35A suppressed clonogenic potential of leukaemia cells in vitro and delayed leukaemogenesis in vivo. We further showed that AML1-ETO9a, MYC and MLL-AF9 all enhanced snoRNA formation. Expression levels of C/D box snoRNAs in AML patients correlated closely with in vivo frequency of leukaemic stem cells. Collectively, these findings indicate that induction of C/D box snoRNA/RNP function constitutes an important pathway in leukaemogenesis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Argiropoulos, B. & Humphries, R. K. Hox genes in hematopoiesis and leukemogenesis. Oncogene 26, 6766–6776 (2007).
Wang, Y. et al. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science 327, 1650–1653 (2010).
Mulloy, J. C. et al. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood 99, 15–23 (2002).
Tonks, A. et al. Expression of AML1-ETO in human myelomonocytic cells selectively inhibits granulocytic differentiation and promotes their self-renewal. Leukemia 18, 1238–1245 (2004).
Yan, M. et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat. Med. 12, 945–949 (2006).
Schessl, C. et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J. Clin. Invest. 115, 2159–2168 (2005).
Müller-Tidow, C. et al. Translocation products in acute myeloid leukemia activate the Wnt signaling pathway in hematopoietic cells. Mol. Cell Biol. 24, 2890–2904 (2004).
Steffen, B. et al. AML1/ETO induces self-renewal in hematopoietic progenitor cells via the Groucho-related amino-terminal AES protein. Blood 117, 4328–4337 (2011).
Hsieh, A. C. et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer cell 17, 249–261 (2010).
Hsieh, A. C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012).
Ruggero, D. & Pandolfi, P. P. Does the ribosome translate cancer? Nat. Rev. Cancer 3, 179–192 (2003).
Barna, M. et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975 (2008).
Signer, R. A., Magee, J. A., Salic, A. & Morrison, S. J. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54 (2014).
Arabi, A. et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7, 303–310 (2005).
Grandori, C. et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 7, 311–318 (2005).
Kiss, T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109, 145–148 (2002).
Sonoshita, M. et al. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell 19, 125–137 (2011).
Wang, W. F. et al. Growth defect in Grg5 null mice is associated with reduced Ihh signaling in growth plates. Dev. Dyn. 224, 79–89 (2002).
Viale, A. et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature 457, 51–56 (2009).
Wang, L. et al. Regulation of AKT signaling by Id1 controls t(8;21) leukemia initiation and progression. Blood 126, 640–650 (2015).
Vangala, R. K. et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood 101, 270–277 (2003).
Pabst, T. et al. AML1-ETO downregulates the granulocytic differentiation factor C/EBPα in t(8;21) myeloid leukemia. Nat. Med. 7, 444–451 (2001).
Reichow, S. L., Hamma, T., Ferre-D’Amare, A. R. & Varani, G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 35, 1452–1464 (2007).
Schwartz, S. et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159, 148–162 (2014).
Baxter-Roshek, J. L., Petrov, A. N. & Dinman, J. D. Optimization of ribosome structure and function by rRNA base modification. PLoS ONE 2, e174 (2007).
van Riggelen, J., Yetil, A. & Felsher, D. W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 10, 301–309 (2010).
Cai, X. et al. Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell 17, 165–177 (2015).
Liu, J., Xu, Y., Stoleru, D. & Salic, A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl Acad. Sci. USA 109, 413–418 (2012).
Calo, E. et al. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 518, 249–253 (2015).
Burel, S. A. et al. Dichotomy of AML1-ETO functions: growth arrest versus block of differentiation. Mol. Cell. Biol. 21, 5577–5590 (2001).
Pabst, C. et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 127, 2018–2027 (2016).
Müller-Tidow, C. et al. Azacitidine in combination with intensive induction chemotherapy in older patients with acute myeloid leukemia: the AML-AZA trial of the study alliance leukemia. Leukemia 30, 555–561 (2016).
Watkins, N. J. et al. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell 16, 789–798 (2004).
Su, H. et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 33, 1348–1358 (2014).
Herter, E. K. et al. snoRNAs are a novel class of biologically relevant Myc targets. BMC Biol. 13, 25–41 (2015).
Sanchez, C. G. et al. Regulation of ribosome biogenesis and protein synthesis controls germline stem cell differentiation. Cell Stem Cell 18, 276–290 (2015).
Basu, A. et al. Requirement of rRNA methylation for 80S ribosome assembly on a cohort of cellular internal ribosome entry sites. Mol. Cell. Biol. 31, 4482–4499 (2011).
Marcel, V. et al. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 24, 318–330 (2013).
Lechman, E. R. et al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell 29, 602–606 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Ulitsky, I. et al. Expander: from expression microarrays to networks and functions. Nat. Protoc. 5, 303–322 (2010).
Oellerich, T. et al. FLT3-ITD and TLR9 use Bruton tyrosine kinase to activate distinct transcriptional programs mediating AML cell survival and proliferation. Blood 125, 1936–1947 (2015).
Kolodziej, S. et al. PADI4 acts as a coactivator of Tal1 by counteracting repressive histone arginine methylation. Nat. Commun. 5, 3995 (2014).
Vinayagam, A. et al. Protein complex-based analysis framework for high-throughput data sets. Sci. Signal. 6, rs5 (2013).
Uren, P. J. et al. Site identification in high-throughput RNA-protein interaction data. Bioinformatics 28, 3013–3020 (2012).
Machnicka, M. A. et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
Belin, S. et al. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS ONE 4, e7147 (2009).
Dong, Z. W. et al. RTL-P: a sensitive approach for detecting sites of 2′-O-methylation in RNA molecules. Nucleic Acids Res. 40, e157 (2012).
Acknowledgements
We thank T. Gridley for providing Aes straight knockout (Aes−/−) mice. We are grateful to B. Edemir for help with project organization and manuscript preparation. We thank M. Scheller-Wendorff for support of the mice experiments. This work was supported by Deutsche Forschungsgemeinschaft (DFG, MU1328/15-1 and MU1328/9-2), the Deutsche Jose Carreras Leukämie-Stiftung e. V. (DJCLS R10/35f), by a Wilhelm-Roux-Program grant (28/43) of the Medical Faculty, Martin-Luther-University Halle-Wittenberg to CMT and the grants of the federal state of Saxony-Anhalt (P40). N.B. is supported by Wilhelm Sander Stiftung (2014.054.1), Deutsche Krebshilfe (70112282) and ‘Innovative Medical Research’ of the University of Münster Medical School (IMF) (121314 and 111501). C.P. is supported by a Max-Eder grant of the Deutsche Krebshilfe (70111531). The laboratory of R.S. is supported by Deutsche Forschungsgemeinschaft (DFG SL27/7-2). The laboratory of W.E.B. is supported by DFG grant EXC 1003. The work of 20 AML samples with LSC frequencies was supported by the Government of Canada through Genome Canada and the Ministère de l’enseignement supérieur, de la recherche, de la science et de la technologie du Québec through Génome Québec to G.S. and J.H. and by grants from the Cancer Research Network of the Fonds de recherche du Québec-Santé to J.H.
Author information
Authors and Affiliations
Contributions
F.Z. designed and conducted the experiments, analysed the data and wrote the manuscript. Y.L. and C.P. made the snoRNA knockouts. M.K., D.M. and S.H. analysed snoRNA-seq data. T.O. and H.S. performed mass spectrometry analyses and analysed the data. C.P. helped with NSG mice experiments. M.-P.G.-C. and R.S. isolated nascent RNA. B.P., C.R., J.P.M., H.-U.K. C.B. and M.D. analysed NGS data. M.M.T. provided the Aesf/f mice. D.F., S.S. and A.R. performed pseudouridylation-sequencing. D.G., C.P., N.B., C.C., S.G., W.E.B. and B.S. discussed the results and advised on research. W.E.B. provided patient samples from the AML-AZA clinical study. J.H. and G.S. provided human AML specimens with determined LSC frequency. C.M.-T. designed and conceptualized the overall research, analysed the data, and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Aes Depletion Inhibits AML1-ETO Induced Self-renewal.
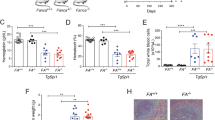
(a,b) FACS analysis of GFP+ transplanted cells in the peripheral blood of recipients, 4 weeks after transplantation. (a) shows a representative FACS plot. (b) shows quantification of the FACS results. (c) White blood cell counts in peripheral blood of recipients, 4 weeks after transplantation. (d) Survival analysis of primary recipients transplanted with AE9a transduced Aes+/+ (n = 12 mice) or Aes−/− (n = 12 mice) bone marrow cells. P = 0.479, log-rank test. (e) Survival of secondary recipients transplanted with AE9a leukemia blast from Aes+/+ or Aes−/− group. Cells form three donors of each group were used for secondary transplantation. N = 12 mice for Aes+/+ group and 10 mice for Aes−/− group, P = 0.004, log-rank test. (f) Experimental scheme for in vitro serial replating assay with AE9a transduced Aes+/+ or Aesf/f murine fetal liver cells. Excision of the floxed Aes alleles was performed by 4-hydroxytamoxifen (4-OHT) treatment either during the first round of replating or after three round of replating. Colonies were counted at each round of replating. (g) Genotype analysis demonstrated the excision of floxed Aes allele. (h) Colony numbers in each round of replating assay as shown in f. Error bars, mean ± s.d. of n = 3 independent experiments. (i) Number of colonies formed by AE9a transformed Aes+/+or Aesf/f fetal liver cells. Error bars, mean ± s.d. of n = 3 independent experiments; ∗∗∗P < 0.001, Student’s t-test. (j) Representative colonies in methylcellulose. Scale bars, 2.5 mm. (k) Western blot shows knockdown of AES by two independent shRNA in Kasumi-1 cells. (l) Colony numbers from 300 indicated leukemic cells with AES knockdown by shRNA, compared to the control (shCtr). Error bars, mean ± s.d. of n = 3 independent experiments. Unprocessed original scans of blots are shown in Supplementary Figure 7. Statistical source data for Supplementary Fig. 1b, c are provided in Supplementary Table 7.
Supplementary Figure 2 AES Regulates SnoRNA/RNPs in Hematopoietic and Leukemia Cells.
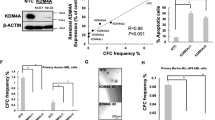
(a) RT-PCR analysis of AE9a target genes in control (shCtr) and AES (shAES) knockdown Kasumi-1 cells. (b) RT-PCR analysis of snoRNA expression to confirm microarray results. GFP positive (AE9a transduced) bone marrow cells with wildtype (Aes+/+) or knockout (Aesf/f) genotype were sorted from recipient mice. cDNA was reverse transcribed from the same RNA used for microarray analysis. Data shown as average of n = 2 biological replicates each group. (c) Quantitative real-time PCR analysis of Aes mRNA in fetal liver cells retrovirally transduced with Aes or empty vector (Control). GAPDH was used for internal control. (d) Colony numbers from fetal liver cells retrovirally transduced with Aes or empty vector. Error Bar, mean ± s.d. n = 3 independent experiments, P = 0.001, Student’s t-test. (e) Comparison of snoRNA expression in Kasumi-1 cells as determined by snoRNA-seq and by qRT-PCR. The x axis shows the relative expression of indicated snoRNAs in AES knockdown cells compared to control cells. (f) RT-PCR analysis of snoRNA expression in control (shCtr) and AES knockdown (shAES) SKNO-1 cells. (g) Scatterplot comparing pseudouridylation ratios at 72 sites in 18S and 28S rRNA in control (shCtr) and AES knockdown (shAES) Kasumi-1 cells. Pseudouridylation were measured after CMC treatment followed by high throughput sequencing, and the plotted Ψ-ratios were calculated as described in ref. 1. Paired Wilcoxon test. (h) Protein expression of small nucleolar ribonucleoprotein (snoRNPs) components Pontin, NOP56, NOP58 and Fibrillarin in AES knockdown (shAES) and control (shCtr) Kasumi-1 cells (Western blot analysis). (i) Forward Scatter size analysis of control (shCtr) and AES knockdown (shAES) Kasumi-1 cells. One of n = 3 independent experiments is shown. (j) OP-Puro incorporation of control (shCtr) and AES knockdown (shAES) Kasumi-1 cells. One of n = 4 independent experiments is shown. Error bars, mean ± s.d. of n = 3 independent experiments unless otherwise indicated. Unprocessed original scans of blots are shown in Supplementary Figure 7. Statistical source data for Supplementary Fig. 2i, j are provided in Supplementary Table 7.
Supplementary Figure 3 AES Interacts with RNA Helicase DDX21 and Multiple SnoRNAs.
(a) Schematic diagram of nascent RNA-Seq. Control (shCtr) and AES knockdown (shAES) Kasumi-1 cells (two replicates for each) were cultured in medium containing 4-thiouridine for 1 h. 4-thiouridine was metabolically incorporated into newly transcribed (nascent) RNA during transcription. Following isolation of total cellular RNA and thiol-specific biotinylation, nascent RNA was separated from total RNA using streptavidin-coated magnetic beads. Paired-end libraries prepared from nascent RNA were sequenced on an Illumina Hiseq2000 platform. (b) Levels of Nascent transcripts (FPKM) of snoRNA host genes in control (shCtr) and AES knockdown cells (shAES). 36 host genes for snoRNAs that are downregulated by AES knockdown were shown. (c) Visualization nascent RNA-seq reads on snoRNA host gene RPL3. (d,e) Gene ontology analysis of downregulated nascent transcripts upon AES knockdown. 2180 transcripts were found to be downregulated in AES knockdown Kasumi-1 cells (FPKMshAES/FPKMshCtr < 0.7 and FPKMshCtr > 1). (f) AES and DDX21 interact in vivo. 293T cells were transfected with V5-tagged AES. Immunoprecipitation was performed with anti-V5 antibody. Western blot analysis was performed with anti-DDX21 or anti-V5 antibody. (g) Western blot indicates DDX21 expression in Kasumi-1 leukemic cells with DDX21 knockdown by shRNA, compared to control shRNA (against luciferase gene, shLUC). (h) RT-PCR analysis of snoRNAs in control and DDX21 knockdown Kasumi-1 cells. Data are represented as mean ± s.d. of n = 3 independent experiments. (i) Kasumi-1 cells were infected with lentivirus expressing control shRNA (shLUC), shRNA against AES or DDX21. Percentage of infected cells (GFP+) was analyzed at the indicated time points. Percentage was normalized to the number at day 1. Error Bar, mean ± s.d. n = 3 independent experiments. (j) Cell cycle analysis of control (shLUC) Kasumi-1 cells and Kasumi-1 knockdown of AES (shAES), DDX21 (shDDX21). Error Bar, mean ± s.d. n = 3 independent experiments. Unprocessed original scans of blots are shown in Supplementary Figure 7. Statistical source data for Supplementary Fig. 3j are provided in Supplementary Table 7.
Supplementary Figure 4 Enrichment of DDX21Interation for ribosome complex.
(a) AES knockdown did not affect transcription from DDX21 bound promoters. Nascent transcripts levels (FPKM) in control and AES knockdown Kasumi-1 cells were used from Nascent RNA-seq as described in Supplementary Fig. 3. DDX21 bound genomic regions were obtained from publically available ChIP-Seq data (GSE56802). Box plots represent fold change (shAES/shCtr, log2 scale) of transcripts originating from promoter regions either bound or not bound by DDX21. As indicated, DDX21-bound gene transcripts were not generally suppressed or enhanced upon loss of AES. The central mark in box plot is mean, with 5/95 percentiles at the whiskers and 25/75 percentiles at the box. (b) DDX21 interaction was enriched for ribosome complex. Mass Spectrometry analysis of SILAC labelled proteins was performed for DDX21 interaction partners at the presence or absence of AES. IP with anti-DDX21 antibody (or IgG control) was performed in protein lysates from Kasumi-1 cells transduced with either shAES or shControl. The ratio of precipitated interaction partners (anti-DDX21/IgG) was analyzed. Circles in red indicate proteins found to interact with DDX21. Proteins labelled in gray are known ribosome complex members that were not identified in our Mass Spectrometry analysis. Gray lines designate protein interactions.
Supplementary Figure 5 Suppression of C/D Box SnoRNAs Delays Leukemogenesis in vivo.
(a) Sanger sequencing of Kasumi-1 CRISPR snoRNA knockout cells. Arrow shows the mutations induced by CRISPR/CAS9 in snoRNA loci. (b) to (c) SnoRNA knockout did not affect host gene expression. SnoRNA host gene RPL13A was analyzed both on mRNA level by RT-PCR (b) and on protein level by Western blot (c) in control and SNORD34 or 35A knockouts. Two single clones for each knockout were checked. (d) SnoRNA knockout did not affect total RNA content. Total RNA from 1 × 105 indicated Kasumi-1cells were eluted in 25 μl RNase-free H2O. Error Bar, mean ± s.d. of n = 5 measurement of 2 independent replicates. (e) Colony numbers formed by MV4-11 CRISPR snoRNA knockout cells, compared to control. (f) Kaplan–Meier survival analysis of NSG mice injected with snoRNA knockout or control MV4-11 cells. N = 12 (Control_gRNA), 7 (SNORD14D_gRNA) and 6 mice (SNORD35_gRNA), log-rank test. (g) Kaplan–Meier survival curve of NSG mice receiving 1 × 104 control (Ctr_gRNA, n = 12 mice) or SNORD43 CRISPR knockout (SNORD43_gRNA, n = 6 mice) MV4-11 cells. Error Bar, means ± s.d. of n = 3 independent experiments unless otherwise indicated. Unprocessed original scans of blots are shown in Supplementary Figure 7.
Supplementary Figure 6 Leukemia oncogenes induce SnoRNA expression.
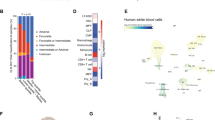
(a) RT-PCR analysis shows expression of snoRNAs in AE9a transduced lin− bone marrow cells, compared to control. (b) and (c) Scatterplot shows snoRNA expression in myc- (b), MLL/AF9-transduced (c) and control (empty vector) lin− bone marrow cells determined by snoRNA-seq. Paired Wilcoxon test. (d) RT-PCR analysis of Aes mRNA expression in lin− bone marrow cells transduce with AE9a, MLL-AF9 or c-Myc, compared to control. (e) and (f) Scatterplot shows snoRNA expression in MLL-AF9 (e) and c-Myc (f) transduced AES wildtype (AES WT) and knockout (AES KO) lin− bone marrow cells determined by snoRNA-seq. Lin− bone marrow cells isolated from ROSA26-Cre/ERT; Aes+/+ or ROSA26-Cre/ERT; Aesf/f were retrovirally transduced with oncogene. Transduced cells were treated with 4-hydroxytamoxifen for three days for Aes deletion. (g) Kaplan–Meier survival analysis of mice injected with MLL-AF9 transduced lin− bone marrow cells isolated from Cre/ERT; Aes+/+ or ROSA26-Cre/ERT; Aesf/f mice. Tamoxifen induction was performed at two weeks after transplantation. N = 7 and 8 mice, log-rank test. (h) Kaplan–Meier survival analysis of mice injected with c-Myc transduced lin− bone marrow cells isolated from AES wildtype (Aes+/+) or straight knockout (Aes−/−) mice. N = 12 mice each group, log-rank test. (i) Cell cycle analysis of U937 cells expressing empty vector (MIG) or AML1-ETO. One of n = 3 independent experiment is shown. (j) Cell cycle distribution of control (MIG) or AML1-ETO-expressing (AE) U937 cells. (k) Scatterplot shows snoRNA expression in control (MIG) or AML-ETO-expressing U937 cells determined by snoRNA-seq. (l) RT-PCR analysis shows expression of snoRNAs in control (MIG) or AML-ETO-expressing (AE) U937 cells. (m) Pseudouridylation ratios were analyzed at 72 sites in 18S and 28S rRNA in human primary AML1-ETO (n = 5) and CD34+ cell samples (n = 5). (n) Heatmap shows significant snoRNAs associated with chemotherapy response. Error bars, mean ± s.d. of n = 3 independent experiments unless otherwise indicated. Statistical source data for Supplementary Fig. 6j are provided in Supplementary Table 7.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1842 kb)
Supplementary Table 1
Supplementary Information (XLSX 40 kb)
Supplementary Table 2
Supplementary Information (XLSX 21 kb)
Supplementary Table 3
Supplementary Information (XLSX 14 kb)
Supplementary Table 4
Supplementary Information (XLSX 12 kb)
Supplementary Table 5
Supplementary Information (XLSX 36 kb)
Supplementary Table 6
Supplementary Information (XLSX 11 kb)
Supplementary Table 7
Supplementary Information (XLSX 50 kb)
Rights and permissions
About this article
Cite this article
Zhou, F., Liu, Y., Rohde, C. et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat Cell Biol 19, 844–855 (2017). https://doi.org/10.1038/ncb3563
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3563
This article is cited by
-
RNA modifications in physiology and disease: towards clinical applications
Nature Reviews Genetics (2024)
-
Small nucleolar RNA Snora73 promotes psoriasis progression by sponging miR-3074-5p and regulating PBX1 expression
Functional & Integrative Genomics (2024)
-
N6-methyladenosine-modified oncofetal lncRNA MIR4435-2HG contributed to stemness features of hepatocellular carcinoma cells by regulating rRNA 2′-O methylation
Cellular & Molecular Biology Letters (2023)
-
RNA modifications in cancer
British Journal of Cancer (2023)
-
LncRNA INHEG promotes glioma stem cell maintenance and tumorigenicity through regulating rRNA 2’-O-methylation
Nature Communications (2023)