Abstract
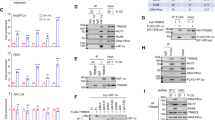
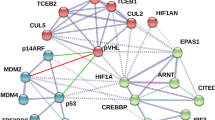
There are three prolyl hydroxylases (PHD1, 2 and 3) that regulate the hypoxia-inducible factors (HIFs), the master transcriptional regulators that respond to changes in intracellular O2 tension1,2. In high O2 tension (normoxia) the PHDs hydroxylate two conserved proline residues on HIF-1α, which leads to binding of the von Hippel–Lindau (VHL) tumour suppressor, the recognition component of a ubiquitin–ligase complex, initiating HIF-1α ubiquitylation and degradation3,4,5,6. However, it is not known whether PHDs and VHL act separately to exert their enzymatic activities on HIF-1α or as a multiprotein complex. Here we show that the tumour suppressor protein LIMD1 (LIM domain-containing protein) acts as a molecular scaffold, simultaneously binding the PHDs and VHL, thereby assembling a PHD–LIMD1–VHL protein complex and creating an enzymatic niche that enables efficient degradation of HIF-1α. Depletion of endogenous LIMD1 increases HIF-1α levels and transcriptional activity in both normoxia and hypoxia. Conversely, LIMD1 expression downregulates HIF-1 transcriptional activity in a manner depending on PHD and 26S proteasome activities. LIMD1 family member proteins Ajuba and WTIP also bind to VHL and PHDs 1 and 3, indicating that these LIM domain-containing proteins represent a previously unrecognized group of hypoxic regulators.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Epstein, A. C. et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 (2001).
Bruick, R. K. & McKnight, S. L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294, 1337–1340 (2001).
Jaakkola, P. et al. Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001).
Ivan, M. et al. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 (2001).
Yu, F., White, S. B., Zhao, Q. & Lee, F. S. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl Acad. Sci. USA 98, 9630–9635 (2001).
Masson, N., Willam, C., Maxwell, P. H., Pugh, C. W. & Ratcliffe, P. J. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 20, 5197–5206 (2001).
Sharp, T. V. et al. LIM domains-containing protein 1 (LIMD1), a tumour suppressor encoded at chromosome 3p21.3, binds pRB and represses E2F-driven transcription. Proc. Natl Acad. Sci. USA 101, 16531–16536 (2004).
Sharp, T. V. et al. The chromosome 3p21.3-encoded gene, LIMD1, is a critical tumour suppressor involved in human lung cancer development. Proc. Natl Acad. Sci. USA 105, 19932–19937 (2008).
Schmeichel, K. L. & Beckerle, M. C. The LIM domain is a modular protein-binding interface. Cell 79, 211–219 (1994).
Kadrmas, J. L. & Beckerle, M. C. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5, 920–931 (2004).
Appelhoff, R. J. et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 279, 38458–38465 (2004).
Berra, E. et al. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 22, 4082–4090 (2003).
Rankin, E. B. & Giaccia, A. J. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 15, 678–685 (2008).
Huggins, C. J. & Andrulis, I. L. Cell cycle regulated phosphorylation of LIMD1 in cell lines and expression in human breast cancers. Cancer Lett. 267, 55–66 (2008).
Ivan, M. et al. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl Acad. Sci. USA 99, 13459–13464 (2002).
Feng, Y. & Longmore, G. D. The LIM protein Ajuba influences interleukin-1-induced NF-κB activation by affecting the assembly and activity of the protein kinase Czeta/p62/TRAF6 signalling complex. Mol. Cell Biol. 25, 4010–4022 (2005).
Iliopoulos, O., Ohh, M. & Kaelin, W. G. Jr pVHL19 is a biologically active product of the von Hippel–Lindau gene arising from internal translation initiation. Proc. Natl Acad. Sci. USA 95, 11661–11666 (1998).
Schoenfeld, A., Davidowitz, E. J. & Burk, R. D. A second major native von Hippel–Lindau gene product, initiated from an internal translation start site, functions as a tumour suppressor. Proc. Natl Acad. Sci. USA 95, 8817–8822 (1998).
Liu, W., Xin, H., Eckert, D. T., Brown, J. A. & Gnarra, J. R. Hypoxia and cell cycle regulation of the von Hippel–Lindau tumour suppressor. Oncogene 30, 21–31 (2011).
Feng, Y. et al. A multifunctional lentiviral-based gene knockdown with concurrent rescue that controls for off-target effects of RNAi. Genom. Proteom. Bioinform. 8, 238–245 (2010).
Chan, D. A., Sutphin, P. D., Yen, S. E. & Giaccia, A. J. Coordinate regulation of the oxygen-dependent degradation domains of hypoxia-inducible factor 1α. Mol. Cell Biol. 25, 6415–6426 (2005).
Stiehl, D. P. et al. Increased prolyl 4-hydroxylase domain proteins compensate for decreased oxygen levels. Evidence for an autoregulatory oxygen-sensing system. J. Biol. Chem. 281, 23482–23491 (2006).
Ginouves, A., Ilc, K., Macias, N., Pouyssegur, J. & Berra, E. PHDs overactivation during chronic hypoxia ‘desensitizes’ HIFα and protects cells from necrosis. Proc. Natl Acad. Sci. USA 105, 4745–4750 (2008).
Tan, M. et al. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1α ubiquitination and degradation. Oncogene 27, 1404–1411 (2008).
Wang, G. L. & Semenza, G. L. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J. Biol. Chem. 268, 21513–21518 (1993).
Berra, E. et al. Signaling angiogenesis via p42/p44 MAP kinase and hypoxia. Biochem. Pharmacol. 60, 1171–1178 (2000).
Minet, E., Michel, G., Mottet, D., Raes, M. & Michiels, C. Transduction pathways involved in Hypoxia-Inducible Factor-1 phosphorylation and activation. Free Radic. Biol. Med. 31, 847–855 (2001).
Richard, D. E., Berra, E., Gothie, E., Roux, D. & Pouyssegur, J. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1α (HIF-1α) and enhance the transcriptional activity of HIF-1. J. Biol. Chem. 274, 32631–32637 (1999).
Huang, L. E., Gu, J., Schau, M. & Bunn, H. F. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin–proteasome pathway. Proc. Natl Acad. Sci. USA 95, 7987–7992 (1998).
Metzen, E. et al. Intracellular localisation of human HIF-1α hydroxylases: implications for oxygen sensing. J. Cell Sci. 116, 1319–1326 (2003).
Lando, D. et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 (2002).
Hubbi, M. E., Luo, W., Baek, J. H. & Semenza, G. L. MCM proteins are negative regulators of hypoxia-inducible factor 1. Mol. Cell 42, 700–712 (2011).
Nakayama, K. & Ronai, Z. Siah: new players in the cellular response to hypoxia. Cell Cycle 3, 1345–1347 (2004).
Barth, S. et al. The peptidyl prolyl cis/trans isomerase FKBP38 determines hypoxia-inducible transcription factor prolyl-4-hydroxylase PHD2 protein stability. Mol. Cell Biol. 27, 3758–3768 (2007).
Barth, S. et al. Hypoxia-inducible factor prolyl-4-hydroxylase PHD2 protein abundance depends on integral membrane anchoring of FKBP38. J. Biol. Chem. 284, 23046–23058 (2009).
Langer, E. M. et al. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev. Cell 14, 424–436 (2008).
Naldini, L. et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272, 263–267 (1996).
Epstein, A. C. et al. C. elegans EGL-9 and mammalian homologs define afamily of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 (2001).
Sharp, T. V. et al. LIM domains-containing protein 1 (LIMD1), a tumour suppressor encoded at chromosome 3p21.3, binds pRB and represses E2F-driven transcription. Proc. Natl Acad. Sci. USA 101, 16531–16536 (2004).
Sharp, T. V. et al. K15 protein of Kaposi’s sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J. Virol. 76, 802–816 (2002).
Beitzinger, M., Peters, L., Zhu, J. Y., Kremmer, E. & Meister, G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 4, 76–84 (2007).
Acknowledgements
We thank R. Layfield, T. Hagen, M. Cockman and S. Kristjansdottir for reagents and technical advice/assistance. K.S.B. is supported by a Biotechnology and Biological Sciences Research Council Doctorate Training Award. V.J. and D.E.F. were supported by funding from the Biotechnology and Biological Sciences Research Council (BB/F006470/1 and BB/I007571/1) awarded to T.V.S.
Author information
Authors and Affiliations
Contributions
D.E.F., K.S.B., V.J., T.M.W. and T.V.S. designed experiments and wrote the paper. M.M., S.C.K.W., D.C-T. and T.E.P. carried out experiments. Y.F., P.J.R., S.I., J.B. and G.D.L. provided reagents. P.J.R. and G.D.L. contributed to experimental design and writing and editing the paper. T.V.S. initiated and managed this investigation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 9466 kb)
Supplementary Table 1
Supplementary Information (XLSX 11 kb)
Supplementary Table 2
Supplementary Information (XLSX 8 kb)
Supplementary Table 3
Supplementary Information (XLSX 9 kb)
Rights and permissions
About this article
Cite this article
Foxler, D., Bridge, K., James, V. et al. The LIMD1 protein bridges an association between the prolyl hydroxylases and VHL to repress HIF-1 activity. Nat Cell Biol 14, 201–208 (2012). https://doi.org/10.1038/ncb2424
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2424
This article is cited by
-
The multifaceted role of EGLN family prolyl hydroxylases in cancer: going beyond HIF regulation
Oncogene (2022)
-
Analysis of genome and methylation changes in Chinese indigenous chickens over time provides insight into species conservation
Communications Biology (2022)
-
HIF-1α switches the functionality of TGF-β signaling via changing the partners of smads to drive glucose metabolic reprogramming in non-small cell lung cancer
Journal of Experimental & Clinical Cancer Research (2021)
-
Targeted therapy for LIMD1-deficient non-small cell lung cancer subtypes
Cell Death & Disease (2021)
-
High nuclear expression of HIF1α, synergizing with inactivation of LIMD1 and VHL, portray worst prognosis among the bladder cancer patients: association with arsenic prevalence
Journal of Cancer Research and Clinical Oncology (2021)