Abstract
As a main regulator of cellular responses to hypoxia, the protein stability of hypoxia-inducible factor (HIF)-1α is strictly controlled by oxygen tension dependent of PHDs-catalyzed protein hydroxylation and pVHL complex-mediated proteasomal degradation. Whether HIF-1α protein stability as well as its activity can be further regulated under hypoxia is not well understood. In this study, we found that OTUB1 augments hypoxia signaling independent of PHDs/VHL and FIH. OTUB1 binds to HIF-1α and depletion of OTUB1 reduces endogenous HIF-1α protein under hypoxia. In addition, OTUB1 inhibits K48-linked polyubiquitination of HIF-1α via its non-canonical inhibition of ubiquitination activity. Furthermore, OTUB1 promotes hypoxia-induced glycolytic reprogramming for cellular metabolic adaptation. These findings define a novel regulation of HIF-1α under hypoxia and demonstrate that OTUB1-mediated HIF-1α stabilization positively regulates HIF-1α transcriptional activity and benefits cellular hypoxia adaptation.
Similar content being viewed by others
Introduction
Hypoxia signaling regulation is the most important cellular process for adapting to physiological oxygen variations, and its dysregulation leads to various pathophysiological dysfunction [1,2,3,4]. The master molecules of the hypoxia signaling are the hypoxia-inducible factors (HIF-1 and HIF-2), which widely controls oxygen responsive genes involved in energy metabolism, angiogenesis, apoptosis, etc. [1, 2, 5, 6]. The α subunit of HIF-1 or HIF-2 is rapidly degraded under normoxia; conversely, this subunit is stabilized when O2-dependent degradation is inhibited under hypoxia [7, 8]. The stability and activity of the α subunit of HIF are regulated by its post-translational modifications, including hydroxylation, acetylation, SUMOylation, phosphorylation, methylation and ubiquitination [9,10,11,12,13,14,15,16,17,18,19,20]. Particularly, the prolyl hydroxylases (PHD1, PHD2, PHD3 or called EglN2, EglN1, EglN3, respectively) hydroxylate a specific residue of the HIF-α subunit (Pro402 and Pro564 on HIF-1α, Pro405 and Pro531 on HIF-2α), to trigger a ubiquitination reaction by E3 ubiquitin ligase Von Hippel-Lindau protein (pVHL) and proteasome-mediated degradation, which represents a canonical hypoxia signaling pathway [21,22,23,24]. While ubiquitination is a powerful HIF regulatory modification, deubiquitination of HIF and its impact on hypoxia signaling is also gotten much more attentions [25, 26].
DUBs are enzymes that can remove ubiquitin chains from proteins [27]. Based on sequences and structure, the DUBs can be divided into seven subgroups, including the ubiquitin-specific proteases (USPs), the ubiquitin carboxyl-terminal hydrolases (UCHs), the otubain/ovarian tumor-domain containing proteins (OTUs), the Machado-Joseph diseases domain superfamily (MJDs), the JAB1/MPN/MOV34 proteases (JAMMs), the monocyte chemotactic protein-induced proteins (MCPIPs), and the novel motif interacting with ubiquitin DUB family (MINDY) [25, 27]. The DUBs play a major role in protecting proteins from proteasomal degradation, thereby affecting various signaling pathway directly or indirectly [28, 29]. Even through it is relatively superficial in understanding the role of deubiquitination in the hypoxia signaling pathway compared to that of ubiquitination, to date, several DUBs, such as USP7, USP8, USP19, USP20, USP28, USP37, UCHL1 and OTUD7B, have been identified to target HIF-α for deubiquitination, resulting in either the enhancement of HIF activity or the suppression of HIF activity [30,31,32,33,34,35,36,37].
OTUB1 (OUT domain-containing ubiquitin aldehyde-binding protein 1) was previously identified as a K48 linkage-specific deubiquitinating enzyme associated with the prevention of protein degradation [38]. OTUB1 has been reported to modulate multiple signaling pathways by deubiquitinating signaling molecules, such as p100, UBE2F1, snail, DEPTOR, YB-1, SMAD2/3, c-IAP, p53, AKT, SOCS1, UBC13, PD-L1, Cyclin E1, MSH2, SLC7A11, TRAF3, and Nur77 [39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57]. Interestingly, the enzymatic activity of OTUB1 can be regulated by factor inhibiting HIF (FIH)-mediated hydroxylation in an oxygen-dependent manner [58, 59]. In fact, FIH is a well-characterized hydroxylase that catalyzes hydroxylation of asparagine residue within HIF-α subunits dependent on oxygen, resulting in the inhibition of HIF-dependent transcription under normoxia [60,61,62,63,64]. This phenomenon provoked us to investigate the impact of OTUB1 on hypoxia signaling.
In this study, we found that OTUB1 augments hypoxia signaling independent of PHDs/VHL and FIH. OTUB1 binds to HIF-1α and depletion of OTUB1 reduces endogenous HIF-1α protein under hypoxia. Furthermore, we found that OTUB1 inhibits K48-linked polyubiquitination of HIF-1α via its non-canonical inhibition of ubiquitination activity.
Materials and methods
Cell line and culture conditions
HEK293T and H1299 cells originally obtained from American Type Culture Collection (ATCC) were cultured in Dulbeccos’ modified Eagle medium (DMEM) (HyClone) with 10% fetal bovine serum (FBS). The cells were grown at 37 °C in a humidified incubator containing 5% CO2. The cells were cultured under hypoxic condition (1% O2, 5% CO2, and balanced with N2) by using the NBS Galaxy 48 R incubator.
Antibodies and chemical reagents
Antibodies including anti-OTUB1 (#3783), anti-HIF-1α (#36169), anti-VHL (#68547), anti-FIH (#4426), anti-Ubiquitin (#3936), anti-K48-linkage Specific Polyubiquitin (#8081), and normal rabbit IgG (#2729) were purchased from Cell Signaling Technology. Anti-ACTB (#AC026) antibody was purchased from ABclonal. Anti-Flag (#F1804) antibody was purchased from Sigma. Anti-HA (#901515) antibody was purchased from Covance. Anti-Myc (#SC-40) antibody was purchased from Santa Cruz Biotechnology. CoCl2 (#C8661), Deferoxamine mesylate salt (DFX) (#D9533), DMOG (#D3695) and MG-132 (#474790) were purchased from Sigma. FG4592 (#S1007) was purchased from Selleck. The cells were treated with DMOG (1 mM) or FG4592 (up to 100 μM) for 6–8 h, and DMSO was used as a control.
Quantitative real-time PCR assay
Total RNAs were extracted using RNAiso Plus (TaKaRa Bio., Beijing, China) following the protocol provided by the manufacturer. cDNAs were synthesized using the Revert Aid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). MonAmpTM SYBR® Green qPCR Mix (high Rox) (Monad Bio., Shanghai, China) was used for quantitative RT–PCR assays (qPCR). The primers for quantitative RT- PCR assays are listed in Supplementary Table S1.
Immunoprecipitation and Western blot
Co-immunoprecipitation and Western blot analysis were performed as described previously [20]. Anti-Flag antibody-conjugated agarose beads (#A2220), anti-HA antibody-conjugated agarose beads (#A2095) and anti-Myc antibody-conjugated agarose beads (#A7470) were purchased from Sigma. Protein G Sepharose (#17–0618–01) was purchased from GE HealthCare Company. The Fuji Film LAS4000 mini-luminescent image analyzer was used to photograph the blots. Image J software (National Institutes of Health) was used to quantify protein levels based on the band density obtained by Western blot analysis.
CRISPR-Cas9 knockout cell lines
To generate H1299 or HEK293T knocked-out cell lines of indicated genes, sgRNA sequence were ligated into Lenti-CRISPRv2 plasmid and then co-transfected with viral packaging plasmids (psPAX2 and pMD2G) into HEK293T cells. Six hours after transfection, medium was changed, and viral supernatant was collected and filtered through 0.45-μm strainer. Targeted cells were infected by viral supernatant and selected by 1 μg/ml puromycin for 2 weeks. The sgRNA sequence targeting VHL was described as previously [65]. The sgRNA sequence targeting OTUB1 is: GGTCCTGCTGAGCCATGA. The sgRNA sequence targeting FIH is: GGGTCGCTCTGACTCAGACG. The sgRNA sequence targeting HIF-1β is: GTCGCCGCTTAATAGCCCTC.
Ubiquitination assay
Ubiquitination assays were followed the protocol described previously with some modifications [66]. Briefly, HEK293T cells were co-transfected with the plasmids expressing Myc-HIF-1α, His-ubiquitin or His-ubiquitin-K48 or His-ubiquitin-K48R, together with Flag-OTUB1 or Flag empty as a control for 24 h and then lysed by denatured buffer (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole), followed by nickel bead purification and immunoblotting with the indicated antibodies.
For ubiquitination assay in OTUB1-deficient or wildtype H1299 cells (OTUB1−/− or OTUB1+/+), the cells were treated with or without MG-132 (20 μM) for 6~8 h, then collected, and lysed with the lysis buffer (100 μl). The supernatants were denatured at 95 °C for 5 min in the presence of 1% SDS. The denatured lysates were diluted with lysis buffer to reduce the concentration of SDS (less than 0.1%). Immunoprecipitation (denature-IP) was conducted using anti-HIF-1α antibody and then subjected to immunoblotting with anti-Ubiquitin or anti-K48-linkage specific polyubiquitin antibody.
Cell proliferation assay
OTUB1-deficient or wildtype H1299 cells (OTUB1−/− or OTUB1+/+) seeded in 96-well plates at 500 cells per well were cultured for indicated days, and CCK-8 assay was employed to determinate the cell growth rate according to the manufacturer’s instructions.
Colony formation assays
OTUB1-deficient or wildtype H1299 cells (OTUB1−/− or OTUB1+/+) seeded in 6-well plates at 2 × 103 cells per well were cultured under hypoxia (1% O2). After 7 days, the colonies were fixed by methanol, stained with crystal violet (0.5% in methanol) and washed with PBS, and then photographed. Colonies of a suitable size were counted based on the images in each well.
Mitochondrial stress test and glycolytic rate test assays
The oxygen consumption rate (OCR) under mitochondrial stress test assay and the proton efflux rate (PER) under glycolytic rate test assay were performed using the Seahorse XFe24 Extracellular Flux Analyzer (Agilent Technologies, Santa Clara, CA, USA). Mitochondrial stress and glycolytic rate test assays were performed using XF Cell Mito Stress Test kit (Agilent Technologies, #103015–100) and XF Glycolytic rate Assay Kit (Agilent Technologies, #103344–100), respectively. The assays were performed according to the manufacturer’s instructions. The H1299 cells (4 × 104 cells/well) were cultured in XF24 cell culture microplate (Agilent Technologies, #102340–100). For mitochondrial stress test assay, oligomycin (1.5 μM), carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP, 2 μM) and antimycin A and rotenone mixture (0.5 μM) were added to cell culture plate for determining mitochondrial respiration including basal respiration, maximal respiration and spare respiratory capacity. For glycolytic rate test assay, antimycin A and rotenone mixture (0.5 μM) and 2-deoxy-D-glucose (50 mM) were added to cell culture plate for determining glycolytic flux including basal glycolysis and compensatory glycolysis.
Glucose uptake assay
Glucose uptake was analyzed directly using the fluorescent glucose analog 2-NBDG. OTUB1-deficient or wildtype H1299 cells (OTUB1−/− or OTUB1+/+) were incubated in glucose-free DMEM medium overnight and then cultured under hypoxia for 6 h. 50 μM 2-NBDG was added into the medium and then the cells were incubated for 1 h at 37 °C in dark, and the amount of 2-NBDG taken up by cells was detected by fluorescence microscopy or assessed by flow cytometry analysis (FACS).
Statistical analysis
GraphPad Prism software (7.0) was used for all statistical analysis. Results with error bars express mean ± SD. Statistical analysis was performed as indicated in figure legends. A P value less than 0.05 was considered significant. Statistical significance is represented as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Results
OTUB1 augments hypoxia signaling
The oxygen-dependent regulation of OTUB1 by FIH provoked us to determinate whether OTUB1 has impact on hypoxia signaling [58, 59]. Initially, we checked the effect of hypoxia on the formation of FIH and OTUB1 heterodimers. In fact, a heterotrimeric complex is indeed formed, which is consisted of FIH and OTUB1 (Fig. S1A). In addition, the formation of this complex is dependent on the hydroxylation of OTUB1 on N22 by FIH and sensitive to hypoxia (Fig. S1B, C), in agreement with the previous report [58, 59]. Furthermore, hypoxia could induce the expression of OTUB1 (Fig. S2A, B). These results implicated that OTUB1 may involve in hypoxia signaling.
Subsequently, we checked the effect of OTUB1 on the expression of hypoxia responsive genes in cells after treated with hypoxia or hypoxia-mimic conditions [1, 20]. Overexpression of OTUB1 in H1299 cells enhanced expression of PGK1, LDHA, VEGF and BNIP3 under hypoxia (1% O2) as revealed by quantitative real-time PCR assays (qPCR) (Fig. 1A–D, S3F) [20, 67]. Similar results were obtained in HEK293T cells (Fig. S3A–E). Consistently, when the cells were treated with either CoCl2 or Deferoxamine mesylate salt (DFX) [68, 69], two hypoxia-mimic conditions, the increased expression of PGK1, LDHA, PDK1 and BNIP3 was also observed in OTUB1-overexpressed H1299 cells (Figs. 1E–L, S3F). Subsequently, we knocked out OTUB1 in H1299 cells via CRISPR/Ca9 technique and examined hypoxia responsive gene expression (Fig. 2A). Under hypoxia (1% O2), expression of PDK1, GLUT1 and LDHA in OTUB1−/− H1299 cells was lower than that in OTUB1+/+ H1299 cells (Fig. 2B–D). In agreement, expression of PDK1, PGK1, GLUT1, LDHA, BNIP3 or VEGF in OTUB1−/− H1299 cells was lower than that in OTUB1+/+ H1299 cells when cells were treatment with either CoCl2 or DFX (Fig. 2E–L). However, reconstitution of OTUB1 in OTUB1−/− H1299 cells promoted expression of PDK1, PGK1, GLUT1 and LDHA compared to reconstitution of empty vector control under hypoxia (1% O2) (Fig. S4A–D, H). These data suggest that OTUB1 enhances HIF transactivity under hypoxia.
A–D qPCR analysis of PGK1 (A), LDHA (B), VEGF (C), and BNIP3 (D) mRNA in H1299 cells transfected with or without Flag-OTUB1 and cultured under normoxia (21% O2) or hypoxia (1% O2) for 24 h. Flag empty vector was used as a control. E–H qPCR analysis of PGK1 (E), LDHA (F), PDK1 (G), and BNIP3 (H) mRNA in H1299 cells transfected with or without Flag-OTUB1 and treated with or without CoCl2 (200 μM) for 8 h. Flag empty vector was used as a control. I–L qPCR analysis of PGK1 (I), LDHA (J), PDK1 (K), and BNIP3 (L) mRNA in H1299 cells transfected with or without Flag-OTUB1 and treated with DFX (150 μM) or DMSO as a control for 8 h. Flag empty vector was used as a control. Two-way ANOVA analysis; Data show mean ± SD; Tukey’s multiple comparisons test; ns, not significant, *Adjusted p < 0.05, **Adjusted p < 0.01, ***Adjusted p < 0.001, ****Adjusted p < 0.0001; Data from 3 independent experiments.
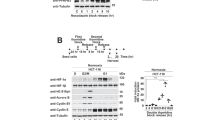
A Western blot analysis of indicated proteins in OTUB1-deficient or wildtype H1299 cells (OTUB1−/− or OTUB1+/+). B–D qPCR analysis of PDK1 (B), GLUT1 (C), and LDHA (D) mRNA in OTUB1-deficient or wildtype H1299 cells (OTUB1−/− or OTUB1+/+) cultured under normoxia (21% O2) or hypoxia (1% O2) for 24 h. Two-way ANOVA analysis; Data show mean ± SD; Tukey’s multiple comparisons test; ns, not significant, *Adjusted p < 0.05, **Adjusted p < 0.01, ***Adjusted p < 0.001, ****Adjusted p < 0.0001; Data from 3 independent experiments. E–H qPCR analysis of PDK1 (E), PGK1 (F), LDHA (G) and BNIP3 (H) mRNA in OTUB1-deficient or wildtype H1299 cells (OTUB1−/− or OTUB1+/+) treated with or without CoCl2 (200 μM) for 8 h. Two-way ANOVA analysis; Data show mean ± SD; Tukey’s multiple comparisons test; ns, not significant, *Adjusted p < 0.05, **Adjusted p < 0.01, ***Adjusted p < 0.001, ****Adjusted p < 0.0001; Data from 3 independent experiments. I–L qPCR analysis of PDK1 (I), GLUT1 (J), VEGF (K), and BNIP3 (L) mRNA in OTUB1-deficient or wildtype H1299 cells (OTUB1−/− or OTUB1+/+) treated with DMSO or DFX (150 μM) for 8 h. Two-way ANOVA analysis; Data show mean ± SD; Tukey’s multiple comparisons test; ns, not significant, *Adjusted p < 0.05, **Adjusted p < 0.01, ***Adjusted p < 0.001, ****Adjusted p < 0.0001; Data from 3 independent experiments.
Given that PHDs-mediated HIF-α hydroxylation represents a main regulatory manner of HIF function [1, 21,22,23], we next sought to determine whether the upregulation of HIF activation by OTUB1 was dependent on PHDs. When H1299 cells were treated with the proline hydroxylase inhibitors, DMOG or FG4592 [70], overexpression of OTUB1 still promoted expression of PDK1, PGK1 or BNIP3 (Fig. 3A–D). In contrast, expression of PDK1, GLUT1, or BNIP3 was lower in OTUB1−/− H1299 cells compared to OTUB1+/+ H1299 cells when either DMOG or FG4592 was added to cells (Fig. S5A–D). Furthermore, we examined whether the upregulation of HIF activation by OTUB1 was dependent on VHL. Knockout of VHL in H1299 cells caused an increased protein level of HIF-1α and an increased mRNA level of PDK1 and PGK1 under normoxia (Fig. 3E, F), indicating that VHL was disrupted efficiently in H1299 cells. Overexpression of OTUB1 enhanced expression of PDK1, PGK1, LDHA, VEGF, and PKM2 in VHL−/− H1299 cells (Fig. 3G). Considering that FIH modulates OTUB1 activity, we subsequently examined whether the upregulation of HIF activation by OTUB1 is dependent on FIH. In FIH-deficient HEK293T cells (Fig. 3H), overexpression of OTUB1 still enhanced expression of PDK1, PGK1 and GLUT1 under hypoxia (1% O2) (Fig. 3I–K). These data suggest that the enhancement of OTUB1 on HIF activity is independent of both PHDs/VHL and FIH.
A, B qPCR analysis of PDK1 (A) and PGK1 (B) mRNA in H1299 cells transfected with or without Flag-OTUB1 and cultured under normoxia (21% O2) for 24 h, followed by treatment with DMSO or DMOG (1 mM) for 8 h. Flag empty vector was used as a control. Two-way ANOVA analysis; Data show mean ± SD; Tukey’s multiple comparisons test; ns, not significant, *Adjusted p < 0.05, **Adjusted p < 0.01, ***Adjusted p < 0.001, ****Adjusted p < 0.0001; Data from 3 independent experiments. C, D qPCR analysis of PDK1 (C) and BNIP3 (D) mRNA in H1299 cells transfected with or without Flag-OTUB1 and cultured under normoxia (21% O2) for 24 h, followed by treatment with DMSO or FG4592 (100 μM) for 8 h. Flag empty vector was used as a control. Two-way ANOVA analysis; Data show mean ± SD; Tukey’s multiple comparisons test; ns, not significant, *Adjusted p < 0.05, **Adjusted p < 0.01, ***Adjusted p < 0.001, ****Adjusted p < 0.0001; Data from 3 independent experiments. E Western blot analysis of indicated proteins in VHL-deficient or wildtype H1299 cells (VHL−/− or VHL+/+) cultured under normoxia (21% O2). F qPCR analysis of PDK1 and PGK1 mRNA in VHL-deficient or wildtype H1299 cells (VHL−/− or VHL+/+) cultured under normoxia (21% O2). Data show mean ± SD; Student’s two tailed t-test; ***p < 0.001; Data from 3 independent experiments. G qPCR analysis of PDK1, PGK1, LDHA, VEGF and PKM2 mRNA in VHL-deficient H1299 cells (VHL−/−) transfected with or without Flag-OTUB1 and cultured under normoxia (21% O2) for 24 h. Flag empty vector was used as a control. Data show mean ± SD; Student’s two tailed t-test; ***p < 0.001; Data from 3 independent experiments. H Western blot analysis of indicated proteins in FIH-deficient or wildtype HEK293T cells (FIH−/− or FIH+/+) cultured under normoxia (21% O2). I–K qPCR analysis of PDK1 (I), PGK1 (J) and GLUT1 (K) mRNA in FIH-deficient HEK293T cells (FIH −/−) transfected with or without Flag-OTUB1 and cultured under normoxia (21% O2) or hypoxia (1% O2) for 24 h. Flag empty vector was used as a control. Two-way ANOVA analysis; Data show mean ± SD; Tukey’s multiple comparisons test; ns, not significant, ****Adjusted p < 0.0001; Data from 3 independent experiments.
OTUB1 interacts with HIF-1α
To determine the effect of OTUB1 on HIF activity mechanistically, we next examined whether OTUB1 interacts with HIF-1α. Ectopic expression of OTUB1 interacted with ectopically expressed HIF-1α in HEK293T cells and vice versa (Fig. 4A, B). In HEK293T cells, endogenous OTUB1 associated with endogenous HIF-1α (Fig. 4C). Domain mapping indicated that ODDD domain, ID and CAD domains of HIF-1α had highest binding capability for OTUB1, but the N-terminal domain (bHLH) did not bind to OTUB1 and was not required for the interaction between OTUB1 and HIF-1α (Fig. 4D, E, S6C, D). In HIF-1β-deficient H1299 cells (HIF-1β−/−), ectopic expression of OTUB1 interacted with ectopically expressed HIF-1α, indicating that hetero-dimerization of HIF-1α and HIF-1β is not required for the interaction between OTUB1 and HIF-1α (Fig. S6A, B). In addition, it appeared that the N-terminal domain (E2/UBD) of OTUB1 did not bind to HIF-1α, while the C-terminal domain of OTUB1 had highest binding capability for HIF-1α (Fig. 4F, G). These data suggest that OTUB1 might argument hypoxia signaling through enhancing HIF-1α function via protein interaction.
A, B Co-immunoprecipitation of Flag-OTUB1 with Myc-HIF-1α and vice versa. HEK293T cells were co-transfected with indicated plasmids and cultured under normoxia (21% O2) for 24 h. Anti-Flag (A) or anti-Myc antibody-conjugated agarose beads (B) were used for immunoprecipitation, and the interaction was detected by immunoblotting with the indicated antibodies. C Endogenous interaction between OTUB1 and HIF-1α. HEK293T cells were cultured under hypoxia (1% O2) for 4 h. Anti-HIF-1α antibody was used for immunoprecipitation, and normal rabbit IgG was used as a control. D Schematic of HIF-1α domains interacted with OTUB1. The interaction is indicated by plus (+) sign. E Co-immunoprecipitation analysis of Flag-OTUB1 with Myc-HIF-1α-truncated mutants. HEK293T cells were co-transfected with the indicated plasmids and cultured under normoxia (21% O2) for 24 h. Anti-Myc antibody-conjugated agarose beads were used for immunoprecipitation, and the interaction was analyzed by immunoblotting with the indicated antibodies. Myc-HIF-1α fragments (HIF-1α-N1, 1–80 aa; HIF-1α-N2, 1–200 aa; HIF-1α-N3, 1–330 aa; HIF-1α-N4, 1–399 aa; HIF-1α-N5, 1–575 aa; HIF-1α-N6, 1–785 aa). F Schematic of OTUB1 domains interacted with HIF-1α. The interaction is indicated by plus (+) sign. G Co-immunoprecipitation analysis of Myc-HIF-1α with Flag-OTUB1-truncated mutants. HEK293T cells were co-transfected with the indicated plasmids and cultured under normoxia (21% O2) for 24 h. Anti-Flag antibody-conjugated agarose beads were used for immunoprecipitation, and the interaction was analyzed by immunoblotting with the indicated antibodies. Flag-OTUB1 fragments (OTUB1-N, 1–80 aa; OTUB1-C, 81–271 aa).
OTUB1 stabilizes HIF-1α by inhibiting its ubiquitination
In analyzing the role of OTUB1 in the hypoxia signaling pathway, we examined the effect of OTUB1 on HIF-1α protein level and found that OTUB1 could upregulate the protein level of HIF-1α (Fig. S4E-G, I). Given that OTUB1 serves as a deubiquitinase, we sought to determine whether OTUB1 can directly affect HIF-1α protein stability. Ectopic expression of OTUB1 in HEK293T cells increased protein level of co-transfected HIF-1α in a dose-dependent manner (Fig. 5A). Similar result was obtained in H1299 cells (Fig. 5B). By contrast, disruption of OTUB1 in H1299 cells caused a reduction of endogenous HIF-1α protein under hypoxia (1% O2) (Fig. 5C). However, OTUB1 had no significant effect on the mRNA level of HIF-1α (Fig. S7A, C).
A Immunoblotting of exogenous Myc-HIF-1α expression in HEK293T cells transfected with an increasing amount of Flag-OTUB1 expression plasmid. B Immunoblotting of exogenous Myc-HIF-1α expression in H1299 cells transfected with an increasing amount of Flag-OTUB1 expression plasmid. C Immunoblotting of endogenous HIF-1α expression in WT or two OTUB1-deficient H1299 cell lines (#1 and #2) cultured under normoxia (21% O2) or hypoxia (1% O2) for 4 h. D Immunoblotting of exogenous HA-HIF-1α-DM (encoding the double mutant of HIF-1α [P402A/P564A]) expression in HEK293T cells transfected with an increasing amount of Flag-OTUB1 expression plasmid. E Immunoblotting of exogenous HA-HIF-1α-TM (encoding the triple mutant of HIF-1α [P402A/P564A/N803A]) expression in HEK293T cells transfected with an increasing amount of Flag-OTUB1 expression plasmid. F Immunoblotting of endogenous HIF-1α expression in WT or two OTUB1-deficient H1299 cell lines treated with DMSO (as a control) or DMOG (1 mM) for 6 h. G Immunoblotting of endogenous HIF-1α expression in WT or OTUB1-deficient H1299 (OTUB1+/+ or OTUB1−/−) cells treated with an increasing amount of FG4592 for 6 h. H Immunoblotting of endogenous HIF-1α expression in WT or OTUB1-deficient H1299 (OTUB1+/+ or OTUB1−/−) cells treated with an increasing time of FG4592 (100 μM). I The relative intensities of HIF-1α in G determined by normalizing the intensities of HIF-1α to the intensities of β-actin. J The relative intensities of HIF-1α in H determined by normalizing the intensities of HIF-1α to the intensities of β-actin. K HIF-1α ubiquitination in HEK293T cells transfected with indicated plasmids for 24 h. L Endogenous HIF-1α ubiquitination in H1299 cells transfected with indicated plasmids and cultured under normoxia (21% O2) for 24 h, followed by MG-132 (20 μM) treatment for 6~8 h. M Endogenous HIF-1α ubiquitination in WT or OTUB1-deficient H1299 (OTUB1+/+ or OTUB1−/−) cells cultured under normoxia (21% O2) and treated with MG-132 (20 μM) for 6~8 h.
Furthermore, ectopic expression of OTUB1 also increased protein levels of co-transfected HIF-1α-DM (in which two Proline residues [P402 and P564] are mutated to Alanine residues) and HIF-1α-TM (in which two Proline residues [P402 and P564] and one Asparagine residue [N803] is mutated to Alanine residue) (Fig. 5D, E), suggesting that the stabilization of HIF-1α protein by OTUB1 might be independent of hydroxylation mediated by PHDs and FIH. In agreement, when DMOG was added, endogenous HIF-1α protein level in OTUB1-knocked out H1299 cells (OTUB1−/− #1 and OTUB1−/− #2) was lower than that in wildtype H1299 cells (OTUB1+/+) (Fig. 5F). Of note, after treated with FG4592 at an increased concentrations or extended time periods, endogenous HIF-1α protein level in OTUB1−/− H1299 cells kept lower than that in OTUB1+/+ H1299 cells (Fig. 5G–J). However, unlike the effect on HIF-1α protein level, OTUB1 did not have a significant effect either on protein levels of VHL and FIH (Fig. S7E, F), or on the mRNA of VHL (Fig. S7B, D).These data indicate that OTUB1 stabilizes HIF-1α independent of HIF-1α hydroxylation mediated by both PHDs and FIH, consistent with the results obtained in the above assays for the effect of OTUB1 on HIF activity.
Besides, MG-132 treatment could block OTUB1’s effect on protein levels of HIF-1α (Fig. S7G). So we examined whether OTUB1 could inhibit ubiquitination of HIF-1α subsequently. Overexpression of OTUB1 in HEK293T cells suppressed ubiquitination of co-transfected HIF-1α (Fig. 5K). Moreover, ectopic expression of OTUB1 in H1299 cells suppressed ubiquitination of endogenous HIF-1α (Fig. 5L). By contrast, knockout of OTUB1 in H1299 (OTUB1−/−) enhanced ubiquitination of HIF-1α (Fig. 5M).
Taken together, these data suggest that OTUB1 stabilizes HIF-1α by inhibiting ubiquitination of HIF-1α.
OTUB1 inhibits K48-linked ubiquitination of HIF-1α via its non-canonical inhibition of ubiquitination activity
OTUB1 was previously identified as a member of the ovarian tumor domain containing a superfamily of proteases that has two distinct activities: canonical enzymatic activity for K48-linked polyubiquitin hydrolysis and non-canonical activity for the formation of E2-repressive complex (Fig. 6A) [49, 50, 71]. To determine which activity is required for OTUB1’s effect on HIF-1α, we constructed two mutants: C91A (defective in canonical deubiquitinase activity) and D88A (defective in binding to E2 enzymes), and examined their effects. In the presence of DMOG, overexpression of OTUB1-D88A in HEK293T cells had no effect on the induction of PKM2 mRNA level, but overexpression of OTUB1-C91A still increased PKM2 mRNA level, similar to that of wildtype OTUB1 (Fig. 6B). Moreover, overexpression of OTUB1-D88A in HEK293T cells did not stabilize co-transfected HIF-1α protein level, but overexpression of OTUB1-C91A still stabilized co-transfected HIF-1α protein level, similar to that of wildtype OTUB (Fig. 6C). However, two mutants could interact with HIF-1α as well (Fig. 6D, E).
A Schematic of the domain architecture of OTUB1, showing its E2-binding/ubiquitin binding domain (UBD) region (E2-UBD), ovarian tumor domain (OTU), catalytic sites and modification sites. The working model shows the canonical enzymatic activity and non-canonical activity mediated by OTUB1. B qPCR analysis of PKM2 mRNA in HEK293T cells transfected with Flag-OTUB1 or its mutants with the treatment of DMSO (as a control) or DMOG (1 mM) for 6 h. Flag empty vector was used as a control. Two-way ANOVA analysis; Data show mean ± SD; Tukey’s multiple comparisons test; ns, not significant, ****Adjusted p < 0.0001; Data from 3 independent experiments. C Immunoblotting of exogenous Myc-HIF-1α expression in HEK293T cells transfected with expression plasmid encoding Flag-OTUB1 or its enzymatically deficient mutants. D Co-immunoprecipitation of Flag-OTUB1-D88A with Myc-HIF-1α. HEK293T cells were co-transfected with indicated plasmids for 24 h. Anti-Flag antibody-conjugated agarose beads were used for immunoprecipitation, and the interaction was detected by immunoblotting with the indicated antibodies. E Co-immunoprecipitation of Flag-OTUB1-C91A with Myc-HIF-1α. HEK293T cells were co-transfected with indicated plasmids for 24 h. Anti-Flag antibody-conjugated agarose beads were used for immunoprecipitation, and the interaction was detected by immunoblotting with the indicated antibodies. F HIF-1α ubiquitination in HEK293T cells transfected with Myc-HIF-1α, His-Ub (His empty vector was used as a control), together with Flag-OTUB1, Flag-OTUB1-D88A or Flag-OTUB1-C91A (Flag empty vector was used as a control) for 24 h. G HIF-1α ubiquitination in HEK293T cells transfected with indicated plasmids for 24 h. H Endogenous HIF-1α ubiquitination in H1299 cells transfected with indicated plasmids for 24 h, followed by MG-132 (20 μM) treatment for 6~8 h. I Endogenous HIF-1α ubiquitination in WT or OTUB1-deficient H1299 (OTUB1+/+ or OTUB1−/−) cells treated with MG-132 (20 μM) for 6~8 h.
Subsequently, we examined the effect of two mutants on ubiquitination of HIF-1α. As shown in Fig. 6F, the ubiquitination inhibition of OTUB1-D88A on HIF-1α is not as dramatic as that of wildtype OTUB1 and OTUB1-C91A (Fig. 6F). As expected, overexpression of OTUB1 reduced K48 linkage-specific ubiquitination of HIF-1α (Fig. 6G). Furthermore, we confirmed that overexpression of OTUB1 in H1299 cells inhibited endogenous K48-linked ubiquitination of HIF-1α (Fig. 6H). By contrast, knockout of OTUB1 in H1299 cells enhanced K48-linked ubiquitination of HIF-1α (Fig. 6I).
Together, these data suggest that OTUB1 inhibits K48-linked ubiquitination of HIF-1α via its non-canonical inhibition of ubiquitination activity.
OTUB1 facilitates hypoxia adaptation
Given the importance of hypoxia signaling in cancer initiation and progression [1, 22, 23], to get insights into the biological function of OTUB1 mediated by its effect on hypoxia signaling, we sought to look into its expression in cancer tissues. Based on the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/), we noticed that expression of OTUB1 in lung cancer tissues was higher than in normal tissues, similar to GLUT1, PDK1 and LDHA (Fig. 7A–D). In addition, the correlation of expression of OTUB1 and GLUT1, LDHA or PGK1 was validated (Figs. 7E, S8A, B). Consistently, OTUB1-deficient H1299 cells (OTUB1−/−) proliferated much slower than OTUB1-intact H1299 cells (OTUB1+/+) (Fig. 7F–H).
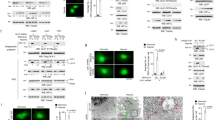
A–D Comparison of GLUT1 (A), PDK1 (B), LDHA (C) and OTUB1 (D) expressions in lung cancer tissues (n = 515) and in the adjacent normal tissues (n = 59). GLUT1 (A), PDK1 (B), LDHA (C) and OTUB1 (D) mRNA levels in tumors were higher than in normal tissues, as determined by the student’s t test. The data were obtained from the Cancer Genome Atlas (TCGA) data (https://cancergenome.nih.gov/) and analyzed by the online tool UALCAN (http://ualcan.path.uab.edu/). E Linear regression of OTUB1 and GLUT1 across the panels of normal and lung cancer samples described in A, D, was generated by the online analysis tool GEPIA (http://gepia.cancer-pku.cn/) based on TCGA data (https://cancergenome.nih.gov/). F Growth curves of WT or OTUB1-deficient H1299 (OTUB1+/+ or OTUB1−/−) cells (n = 5) cultured for the indicated days by CCK-8 assay. G, H Colony formation of WT or OTUB1-deficient H1299 (OTUB1+/+ or OTUB1−/−) cells (n = 3) cultured under hypoxia for the indicated days. I, J Oxygen consumption rate (OCR) changes in wildtype (WT) or OTUB1-deficient H1299 (OTUB1+/+ or OTUB1−/−) cells (n = 5) under normoxia (Nor) or hypoxia (Hyp) measured by Seahorse XFe24 Extracellular Flux Analyzer (I). Statistics of basal respiration, maximal respiration, and spare respiratory capacity were presented in J. K, L Proton efflux rate (PER) changes in WT or OTUB1-deficient H1299 (OTUB1+/+ or OTUB1−/−) cells (n = 5) under normoxia (Nor) or hypoxia (Hyp) measured by Seahorse XFe24 Extracellular Flux Analyzer. Statistics of basal glycolysis and compensatory glycolysis were presented in L. M–O Glucose uptake in WT or OTUB1-deficient H1299 (OTUB1+/+ or OTUB1−/−) cells (n = 3) under normoxia or hypoxia analyzed using fluorescent glucose analog 2-NBDG and detected by fluorescence microscopy (M), and flow cytometry analysis (N, O).
It has been well-established that HIF-1 is a master regulator for metabolic adaptation under hypoxia [72,73,74]. Subsequently, we compared the oxygen consumption rate between wildtype (OTUB1+/+) and OTUB1-deficient H1299 (OTUB1−/−) cells under normoxia or hypoxia based on mitochondrial stress test. Under hypoxia, the maximal respiration and spare respiratory capacity of OTUB1−/− H1299 cells was higher than that of OTUB1+/+H1299 cells (Fig. 7I–J). We next determined the effect of OTUB1 on glycolysis via the proton efflux rate measurement. Loss of OTUB1 in H1299 cells led to decreased compensatory glycolysis significantly compared to wildtype H1299 cells under hypoxia (Fig. 7K, L). Moreover, Loss of OTUB1 in H1299 cells diminished glucose uptake (Fig. 7M–O).
Collectively, these data suggest that OTUB1 facilitates hypoxia adaptation, which might benefit cancer progression.
In order to determine whether the formation of FIH and OTUB1 heterodimers could affect OTUB1’s regulation on hypoxia signaling, we used FG4592 to stabilize HIF-1α under normoxia. Upon FG4592 treatment, FIH had no obvious effect on HIF-1α’s protein level, while OTUB1 could stabilize HIF-1α. However, FIH and OTUB1 co-expression could eliminate the stabilization of HIF-1α by OTUB1 (Fig. S9A, B).
Based on the above observations, a working model for OTUB1 in the regulation of hypoxia signaling is proposed (Fig. 8).
Under normoxia, OTUB1 forms a complex with FIH dependent of oxygen and loses its non-canonical inhibition of ubiquitination activity. Under hypoxia, OTUB1 is dissociated from FIH, which inhibits K48-linked polyubiquitination of HIF-1α. As a result, HIF-1α is stabilized and transactivates downstream genes.
Discussion
The regulation of HIF-1α protein stability under normoxia and hypoxia is well-defined [1]. Under normoxia, PHDs/VHL system keeps HIF-1α protein at almost undetectable level, but, under hypoxia, because PHDs lose enzymatic activity, HIF-1α protein is stabilized and acts its function [1, 75, 76]. Even though the regulation of HIF-1α protein stability by PHDs/VHL accounts for a major regulatory manner for hypoxia signaling, the factors other than PHDs/VHL might also be involved in the regulation of HIF-1α protein stability, particularly some deubiquitinases [25]. Therefore, some efforts have been taken to identify deubiquitinases for regulating HIF [25, 31, 32, 35, 36]. Actually, whether HIF-1α protein stability could be regulated under hypoxia is still a puzzle. In this study, we identified that OTUB1 is involved in regulating HIF-1α activity independent of PHDs/VHL and FIH, and disruption of OTUB1 reduces HIF-1α protein under hypoxia. However, the effect of OTUB1 on HIF-1α function under normoxia conditions was not as consistent as that under hypoxia-mimicking conditions and for that reason the effect of OTUB1 on HIF-1α function in normoxic conditions was not studied. Given that the non-canonical activity of OTUB1 is suppressed by FIH under normoxia [59] and OTUB1 regulates HIF-1α via its non-canonical inhibition of ubiquitination, OTUB1 might contribute to the stabilization of HIF-1α protein under hypoxia. In fact, in VHL-deficient tumors, HIF-1α protein level keeps at very high level, which is considered to be a major cause of cancer progression [75, 76] [36]. Here, we find that OTUB1 is involved in HIF-1α stabilization independent of VHL, so, OTUB1 might be a therapeutic target for the treatment of VHL-deficient tumors.
Even though several deubiquitinases of HIF-α have been identified so far, actually, how they act roles for regulating HIF activity under hypoxia is still not well-understood [25, 26, 31, 35, 36]. Here, inferred from the modulation of OTUB1 by FIH, an important regulator of HIF pathway, we further identified when FIH loses its suppressive role on the non-canonical inhibition ubiquitination activity of OTUB1 under hypoxia, OTUB1 enhances HIF-1α activity, uncovering a connection among FIH/OTUB1/HIF-1α. This finding provides evidence for supporting multiple regulatory manners in hypoxia signaling pathway.
In this study, for the first time, we found that OTUB1, is involved in the regulation of hypoxia signaling. Given that importance of hypoxia signaling in metabolism, cancer progression, and other multiple physiological processes, to investigate the role and the underlying mechanism of OTUB1 in these processes will open a new window for understanding the physiological relevance of OTUB1.
Data availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by Wuhan Xiao.
References
Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol. 2014;9:47–71.
Semenza GL. Hypoxia-inducible factor 1 and cardiovascular disease. Annu Rev Physiol. 2014;76:39–56.
Walshe TE, D’Amore PA. The role of hypoxia in vascular injury and repair. Annu Rev Pathol. 2008;3:615–43.
Lee FS, Percy MJ. The HIF pathway and erythrocytosis. Annu Rev Pathol. 2011;6:165–92.
Wheaton WW, Chandel NS. Hypoxia. 2. Hypoxia regulates cellular metabolism. Am J Physiol Cell Physiol. 2011;300:C385–93.
Harris AL. Hypoxia-a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47.
Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–4.
Corrado C, Fontana S. Hypoxia and HIF signaling: one axis with divergent effects. Int J Mol Sci. 2020;21:16.
Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–78.
Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, et al. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science 2009;324:1289–93.
Bae SH, Jeong JW, Park JA, Kim SH, Bae MK, Choi SJ, et al. Sumoylation increases HIF-1alpha stability and its transcriptional activity. Biochem Biophys Res Commun. 2004;324:394–400.
Berta MA, Mazure N, Hattab M, Pouyssegur J, Brahimi-Horn MC. SUMOylation of hypoxia-inducible factor-1alpha reduces its transcriptional activity. Biochem Biophys Res Commun. 2007;360:646–52.
Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 2007;131:584–95.
Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK, et al. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell 2007;131:309–23.
Mylonis I, Chachami G, Samiotaki M, Panayotou G, Paraskeva E, Kalousi A, et al. Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxia-inducible factor-1alpha. J Biol Chem. 2006;281:33095–106.
Warfel NA, Dolloff NG, Dicker DT, Malysz J, El-Deiry WS. CDK1 stabilizes HIF-1alpha via direct phosphorylation of Ser668 to promote tumor growth. Cell Cycle. 2013;12:3689–701.
Kalousi A, Mylonis I, Politou AS, Chachami G, Paraskeva E, Simos G. Casein kinase 1 regulates human hypoxia-inducible factor HIF-1. J Cell Sci. 2010;123:2976–86.
Xu D, Yao Y, Lu L, Costa M, Dai W. Plk3 functions as an essential component of the hypoxia regulatory pathway by direct phosphorylation of HIF-1alpha. J Biol Chem. 2010;285:38944–50.
Geng H, Harvey CT, Pittsenbarger J, Liu Q, Beer TM, Xue C, et al. HDAC4 protein regulates HIF1alpha protein lysine acetylation and cancer cell response to hypoxia. J Biol Chem. 2011;286:38095–102.
Liu X, Chen Z, Xu C, Leng X, Cao H, Ouyang G, et al. Repression of hypoxia-inducible factor alpha signaling by Set7-mediated methylation. Nucleic Acids Res. 2015;43:5081–98.
Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–28.
Kaelin WG. Von Hippel-Lindau disease. Annu Rev Pathol. 2007;2:145–73.
Kaelin WG Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–73.
Yang G, Shi R, Zhang Q. Hypoxia and oxygen-sensing signaling in gene regulation and cancer progression. Int J Mol Sci. 2020;21:8162.
Mennerich D, Kubaichuk K, Kietzmann T. DUBs, Hypoxia, and Cancer. Trends Cancer. 2019;5:632–53.
Kubaichuk K, Kietzmann T. Involvement of E3 ligases and deubiquitinases in the control of HIF-alpha subunit abundance. Cells 2019;8:598.
Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–92.
Harrigan JA, Jacq X, Martin NM, Jackson SP. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat Rev Drug Disco. 2018;17:57–78.
Kim SY, Baek KH. TGF-beta signaling pathway mediated by deubiquitinating enzymes. Cell Mol Life Sci. 2019;76:653–65.
Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6:373–8.
Troilo A, Alexander I, Muehl S, Jaramillo D, Knobeloch KP, Krek W. HIF1alpha deubiquitination by USP8 is essential for ciliogenesis in normoxia. EMBO Rep. 2014;15:77–85.
Wu HT, Kuo YC, Hung JJ, Huang CH, Chen WY, Chou TY, et al. K63-polyubiquitinated HAUSP deubiquitinates HIF-1alpha and dictates H3K56 acetylation promoting hypoxia-induced tumour progression. Nat Commun. 2016;7:13644.
Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell 2004;117:941–52.
Flugel D, Gorlach A, Kietzmann T. GSK-3beta regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1alpha. Blood 2012;119:1292–301.
Goto Y, Zeng L, Yeom CJ, Zhu Y, Morinibu A, Shinomiya K, et al. UCHL1 provides diagnostic and antimetastatic strategies due to its deubiquitinating effect on HIF-1alpha. Nat Commun. 2015;6:6153.
Hong K, Hu L, Liu X, Simon JM, Ptacek TS, Zheng X, et al. USP37 promotes deubiquitination of HIF2alpha in kidney cancer. Proc Natl Acad Sci USA. 2020;117:13023–32.
Bremm A, Moniz S, Mader J, Rocha S, Komander D. Cezanne (OTUD7B) regulates HIF-1alpha homeostasis in a proteasome-independent manner. EMBO Rep. 2014;15:1268–77.
Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, et al. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol. 2009;386:1011–23.
Li Y, Yang JY, Xie X, Jie Z, Zhang L, Shi J, et al. Preventing abnormal NF-kappaB activation and autoimmunity by Otub1-mediated p100 stabilization. Cell Res. 2019;29:474–85.
Pasupala N, Morrow ME, Que LT, Malynn BA, Ma A, Wolberger C. OTUB1 non-catalytically stabilizes the E2 ubiquitin-conjugating enzyme UBE2E1 by preventing its autoubiquitination. J Biol Chem. 2018;293:18285–95.
Zhou H, Liu Y, Zhu R, Ding F, Cao X, Lin D, et al. OTUB1 promotes esophageal squamous cell carcinoma metastasis through modulating Snail stability. Oncogene 2018;37:3356–68.
Zhao L, Wang X, Yu Y, Deng L, Chen L, Peng X, et al. OTUB1 protein suppresses mTOR complex 1 (mTORC1) activity by deubiquitinating the mTORC1 inhibitor DEPTOR. J Biol Chem. 2018;293:4883–92.
Dong W, Wang H, Shahzad K, Bock F, Al-Dabet MM, Ranjan S, et al. Activated protein C ameliorates renal ischemia-reperfusion injury by restricting Y-box binding protein-1 ubiquitination. J Am Soc Nephrol. 2015;26:2789–99.
Herhaus L, Al-Salihi M, Macartney T, Weidlich S, Sapkota GP. OTUB1 enhances TGFbeta signalling by inhibiting the ubiquitylation and degradation of active SMAD2/3. Nat Commun. 2013;4:2519.
Goncharov T, Niessen K, de Almagro MC, Izrael-Tomasevic A, Fedorova AV, Varfolomeev E, et al. OTUB1 modulates c-IAP1 stability to regulate signalling pathways. EMBO J. 2013;32:1103–14.
Sun XX, Dai MS. Deubiquitinating enzyme regulation of the p53 pathway: a lesson from Otub1. World J Biol Chem. 2014;5:75–84.
Zhou X, Yu J, Cheng X, Zhao B, Manyam GC, Zhang L, et al. The deubiquitinase Otub1 controls the activation of CD8(+) T cells and NK cells by regulating IL-15-mediated priming. Nat Immunol. 2019;20:879–89.
Wang X, Mulas F, Yi W, Brunn A, Nishanth G, Just S, et al. OTUB1 inhibits CNS autoimmunity by preventing IFN-gamma-induced hyperactivation of astrocytes. EMBO J. 2019;38:e100947.
Nakada S, Tai I, Panier S, Al-Hakim A, Iemura S, Juang YC, et al. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 2010;466:941–6.
Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 2012;483:618–22.
Mulas F, Wang X, Song S, Nishanth G, Yi W, Brunn A, et al. The deubiquitinase OTUB1 augments NF-kappaB-dependent immune responses in dendritic cells in infection and inflammation by stabilizing UBC13. Cell Mol Immunol. 2021;18:1512–27.
Zhu D, Xu R, Huang X, Tang Z, Tian Y, Zhang J, et al. Deubiquitinating enzyme OTUB1 promotes cancer cell immunosuppression via preventing ER-associated degradation of immune checkpoint protein PD-L1. Cell Death Differ. 2021;28:1773–89.
Liao Y, Wu N, Wang K, Wang M, Wang Y, Gao J, et al. OTUB1 promotes progression and proliferation of prostate cancer via deubiquitinating and stabling cyclin E1. Front Cell Dev Biol. 2020;8:617758.
Wu Q, Huang Y, Gu L, Chang Z, Li GM. OTUB1 stabilizes mismatch repair protein MSH2 by blocking ubiquitination. J Biol Chem. 2021;296:100466.
Liu T, Jiang L, Tavana O, Gu W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 2019;79:1913–24.
Xie M, Yin Y, Chen L, Yin A, Liu Y, Liu Y, et al. Scavenger receptor A impairs interferon response to HBV infection by limiting TRAF3 ubiquitination through recruiting OTUB1. FEBS J. 2020;287:310–24.
Pei HZ, Huang B, Chang HW, Baek SH. Ovarian tumor domain-containing ubiquitin aldehyde binding protein 1 inhibits inflammation by regulating Nur77 stability. Cell Signal. 2019;59:85–95.
Scholz CC, Rodriguez J, Pickel C, Burr S, Fabrizio JA, Nolan KA, et al. FIH regulates cellular metabolism through hydroxylation of the deubiquitinase OTUB1. PLoS Biol. 2016;14:e1002347.
Pickel C, Gunter J, Ruiz-Serrano A, Spielmann P, Fabrizio JA, Wolski W, et al. Oxygen-dependent bond formation with FIH regulates the activity of the client protein OTUB1. Redox Biol. 2019;26:101265.
Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–71.
Dann CE 3rd, Bruick RK, Deisenhofer J. Structure of factor-inhibiting hypoxia-inducible factor 1: an asparaginyl hydroxylase involved in the hypoxic response pathway. Proc Natl Acad Sci USA. 2002;99:15351–6.
Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–86.
Metzen E, Ratcliffe PJ. HIF hydroxylation and cellular oxygen sensing. Biol Chem. 2004;385:223–30.
Coleman ML, Ratcliffe PJ. Signalling cross talk of the HIF system: involvement of the FIH protein. Curr Pharm Des. 2009;15:3904–7.
Zhang J, Wu T, Simon J, Takada M, Saito R, Fan C, et al. VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science 2018;361:290–5.
Zhu J, Li X, Cai X, Zha H, Zhou Z, Sun X, et al. Arginine monomethylation by PRMT7 controls MAVS-mediated antiviral innate immunity. Mol Cell. 2021;81:3171–86.e8.
Wang J, Zhang D, Du J, Zhou C, Li Z, Liu X, et al. Tet1 facilitates hypoxia tolerance by stabilizing the HIF-alpha proteins independent of its methylcytosine dioxygenase activity. Nucleic Acids Res. 2017;45:12700–14.
Wang GL, Semenza GL. Desferrioxamine induces erythropoietin gene expression and hypoxia-inducible factor 1 DNA-binding activity: implications for models of hypoxia signal transduction. Blood 1993;82:3610–5.
An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature 1998;392:405–8.
Sulser P, Pickel C, Gunter J, Leissing TM, Crean D, Schofield CJ, et al. HIF hydroxylase inhibitors decrease cellular oxygen consumption depending on their selectivity. FASEB J. 2020;34:2344–58.
Jahan AS, Biquand E, Munoz-Moreno R, Le Quang A, Mok CK, Wong HH, et al. OTUB1 is a key regulator of RIG-I-dependent immune signaling and is targeted for proteasomal degradation by influenza A NS1. Cell Rep. 2020;30:1570–84 e6.
Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer. 2008;8:705–13.
Semenza GL. Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb Symp Quant Biol. 2011;76:347–53.
Abu-Remaileh M, Aqeilan RI. Tumor suppressor WWOX regulates glucose metabolism via HIF1 alpha modulation. Cell Death Differ. 2014;21:1805–14.
Kaelin WG Jr. The VHL tumor suppressor gene: insights into oxygen sensing and cancer. Trans Am Clin Climatol Assoc. 2017;128:298–307.
Kaelin WG Jr. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–82.
Acknowledgements
We are grateful to Drs Peter J. Ratcliffe, William Kaelin, Amato Giaccia, Eric Huang, Navdeep Chandel, Bo Zhong, Lingqiang Zhang for the generous gifts of reagents.
Funding
The Strategic Priority Research Program of the Chinese Academy of Sciences [XDA24010308 to WX]; NSFC [31830101 and 31721005 to WX]; the National Key Research and Development Program of China [2018YFD0900602, to WX]. Funding for open access charge: the Strategic Priority Research Program of the Chinese Academy of Sciences [XDA24010308 to WX].
Author information
Authors and Affiliations
Contributions
XL, HD, WL, and WX designed the projects and analyzed the data. XL and HD performed the experiments. JT, ZW, CZ, XC, FR, XC, XS, SJ, and GO helped with the experiments or provided reagents. XL, HD, WL, and WX prepared the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Dr. Ivano Amelio
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, X., Deng, H., Tang, J. et al. OTUB1 augments hypoxia signaling via its non-canonical ubiquitination inhibition of HIF-1α during hypoxia adaptation. Cell Death Dis 13, 560 (2022). https://doi.org/10.1038/s41419-022-05008-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-022-05008-z
This article is cited by
-
OTUB1 accelerates hepatocellular carcinoma by stabilizing RACK1 via its non-canonical ubiquitination
Cellular Oncology (2024)