Abstract
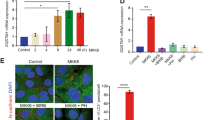
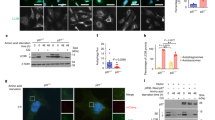
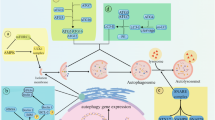
Multiple cellular stressors, including activation of the tumour suppressor p53, can stimulate autophagy. Here we show that deletion, depletion or inhibition of p53 can induce autophagy in human, mouse and nematode cells subjected to knockout, knockdown or pharmacological inhibition of p53. Enhanced autophagy improved the survival of p53-deficient cancer cells under conditions of hypoxia and nutrient depletion, allowing them to maintain high ATP levels. Inhibition of p53 led to autophagy in enucleated cells, and cytoplasmic, not nuclear, p53 was able to repress the enhanced autophagy of p53−/− cells. Many different inducers of autophagy (for example, starvation, rapamycin and toxins affecting the endoplasmic reticulum) stimulated proteasome-mediated degradation of p53 through a pathway relying on the E3 ubiquitin ligase HDM2. Inhibition of p53 degradation prevented the activation of autophagy in several cell lines, in response to several distinct stimuli. These results provide evidence of a key signalling pathway that links autophagy to the cancer-associated dysregulation of p53.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Levine, B. & Kroemer, G. Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008).
Levine, B. & Yuan, J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 115, 2679–2688 (2005).
Degenhardt, K. et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10, 51–64 (2006).
Qu, X. et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946 (2007).
Matthew, R., Karantza-Wadsworth, V. & White, E. Role of autophagy in cancer. Nature Rev. Cancer 7, 961–967 (2007).
Kroemer, G. & Jaattela, M. Lysosomes and autophagy in cell death control. Nature Rev. Cancer 5, 886–897 (2005).
Boya, P. et al. Inhibition of macroautophagy triggers apoptosis. Mol. Cell Biol. 25, 1025–1040 (2005).
Maiuri, C., Zalckvar, E., Kimchi, A. & Kroemer, G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nature Rev. Mol. Cell Biol. 8, 741–752 (2007).
Vousden, K. H. & Lane, D. P. p53 in health and disease. Nature Rev. Mol. Cell Biol. 8, 275–283 (2007).
Feng, Z., Zhang, H., Levine, A. J. & Jin, S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl Acad. Sci. USA 102, 8204–8209 (2005).
Martins, C. P., Brown-Swigart, L. & Evan, G. I. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 127, 1323–1334 (2006).
Xue, W. et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007).
Amaravadi, R. K. et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest. 117, 326–336 (2007).
Crighton, D. et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126, 121–134 (2006).
Bunz, F. et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J. Clin. Invest. 104, 263–269 (1999).
Komarov, P. G. et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science 285, 1733–1737 (1999).
Klionsky, D. J. & al., e. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 (2008).
Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T. & Ohsumi, Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111 (2004).
Criollo, A. et al. Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ. 14, 1029–1039 (2007).
Liang, X. H. et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 (1999).
Mizushima, N. & Yoshimori, T. How to Interpret LC3 Immunoblotting. Autophagy 3 (2007).
Meley, D. et al. AMP-activated protein kinase and the regulation of autophagic proteolysis. J. Biol. Chem. 281, 34870–34879 (2006).
Faivre, S., Kroemer, G. & Raymond, E. Current development of mTOR inhibitors as anticancer agents. Nature Rev. Drug Discov. 5, 671–688 (2006).
Lum, J. J., DeBerardinis, R. J. & Thompson, C. B. Autophagy in metazoans: cell survival in the land of plenty. Natue Rev. Mol. Cell Biol. 6, 439–448 (2005).
Talos, F., Petrenko, O., Mena, P. & Moll, U. M. Mitochondrially targeted p53 has tumor suppressor activities in vivo. Cancer Res. 65, 9971–9981 (2005).
Baker, S. J. et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244, 217–221 (1989).
Tomita, Y. et al. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J. Biol. Chem. 281, 8600–8606 (2006).
Vater, C. A., Bartle, L. M., Dionne, C. A., Littlewood, T. D. & Goldmacher, V. S. Induction of apoptosis by tamoxifen-activation of a p53-estrogen receptor fusion protein expressed in E1A and T24 H-ras transformed p53−/− mouse embryo fibroblasts. Oncogene 13, 739–748 (1996).
Ogata, M. et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol. Cell Biol. 26, 9220–9231 (2006).
Qu, L. et al. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes Dev. 18, 261–277 (2004).
Pluquet, O., Qu, L. K., Baltzis, D. & Koromilas, A. E. Endoplasmic reticulum stress accelerates p53 degradation by the cooperative actions of Hdm2 and glycogen synthase kinase 3β. Mol. Cell Biol. 25, 9392–9405 (2005).
Yorimitsu, T., Nair, U., Yang, Z. & Klionsky, D. J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 281, 30299–30304 (2006).
Vassilev, L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004).
Issaeva, N. et al. Small molecule RITA binds to p53, blocks p53–HDM-2 interaction and activates p53 function in tumors. Nature Med. 10, 1321–1328 (2004).
Kubbutat, M. H., Jones, S. N. & Vousden, K. H. Regulation of p53 stability by Mdm2. Nature 387, 299–303 (1997).
Mihara, M. et al. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11, 577–590 (2003).
Chipuk, J. E., Maurer, U., Green, D. R. & Schuler, M. Pharmacologic activation of p53 elicits Bax-dependent apoptosis in the absence of transcription. Cancer Cell 4, 371–381 (2003).
Chipuk, J. E. et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303, 1010–1014 (2004).
Pattingre, S. et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122, 927–939 (2005).
Leu, J. I., Dumont, P., Hafey, M., Murphy, M. E. & George, D. L. Mitochondrial p53 activates Bak and causes disruption of a Bak–Mcl1 complex. Nature Cell Biol. 6, 443–450 (2004).
Maiuri, M. C. et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 26, 2527–2539 (2007).
Feng, Z. et al. The regulation of AMPK β1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1–AKT–mTOR pathways. Cancer Res. 67, 3043–3053 (2007).
Fabbro, M. & Henderson, B. R. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp. Cell Res. 282, 59–69 (2003).
Toledo, F. & Wahl, G. M. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nature Rev. Cancer 6, 909–923 (2006).
Lim, Y. P. et al. The p53 knowledgebase: an integrated information resource for p53 research. Oncogene 26, 1517–1521 (2007).
Jones, R. G. et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18, 283–293 (2005).
Rubinsztein, D. C., Gestwicki, J. E., Murphy, L. O. & Klionsky, D. J. Potential therapeutic applications of autophagy. Nature Rev. Drug Discov. 6, 304–312 (2007).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Brahimi-Horn, M. C., Chiche, J. & Pouyssegur, J. Hypoxia signalling controls metabolic demand. Curr. Opin. Cell Biol. 19, 223–229 (2007).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–572 8 (2000).
Acknowledgements
We thank N. Mizushima for GFP–LC3-transgenic mice, Levine and B. Vogelstein for cell lines, J. Chipuk, C. G. Maki, M. Oren and K. Vousden for mutant p53 plasmids, E. Zaharioudaki, B. Gardie-Capdeville, C. Ladrou, M.R. Duchen, A. Jalil, F. Fanelli and A. Petrini for expert assistance. The cep-1(gk138) mutant strain (TJ1) was provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). N.T. is funded by EMBO and the EU Sixth Framework Programme. F.C. is funded by Fondazione Telethon, AIRC, Compagnia di San Paolo and the Italian Ministry of Health. E.T. is a recipient of a PhD fellowship from Institut National contre le Cancer (INCa). G.K. is supported by Ligue Nationale contre le Cancer, Agence Nationale pour la Recherche, Cancéropôle Ile-de-France, INCa, Fondation pour la Recherche Médicale, and European Union (Active p53, Apo-Sys, ApopTrain, ChemoRes, TransDeath, RIGHT).
Author information
Authors and Affiliations
Contributions
E.T., M.C.M., L.G. and I.V. conducted experiments, prepared figures and analysed data; M.D.-M., M.D.'A., A.C., E.M., C.Z., F.H., U.N., C.S., P.P., J.M.V, R.C., F.M., P.P.B, G.S., G.P., K.B., N.T., P.C. and F.C. performed experiments; E.T. and G.K. planned the project; G.K. supervised the project and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementay Figures S1, S2, S3, S4, S5, S6 (PDF 1736 kb)
Rights and permissions
About this article
Cite this article
Tasdemir, E., Maiuri, M., Galluzzi, L. et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 10, 676–687 (2008). https://doi.org/10.1038/ncb1730
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1730
This article is cited by
-
Improvement of ACK1-targeted therapy efficacy in lung adenocarcinoma using chloroquine or bafilomycin A1
Molecular Medicine (2023)
-
Protein post-translational modifications in the regulation of cancer hallmarks
Cancer Gene Therapy (2023)
-
Transcriptional regulation of autophagy and its implications in human disease
Cell Death & Differentiation (2023)
-
Sirtuin 4 activates autophagy and inhibits tumorigenesis by upregulating the p53 signaling pathway
Cell Death & Differentiation (2023)
-
Boesenbergia rotunda displayed anti-inflammatory, antioxidant and anti-apoptotic efficacy in doxorubicin‐induced cardiotoxicity in rats
Scientific Reports (2023)