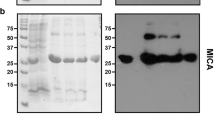
Abstract
Overexpression of proteins in Escherichia coli at low temperature improves their solubility and stability1,2. Here, we apply the unique features of the cspA gene to develop a series of expression vectors, termed pCold vectors, that drive the high expression of cloned genes upon induction by cold-shock. Several proteins were produced with very high yields, including E. coli EnvZ ATP-binding domain (EnvZ-B) and Xenopus laevis calmodulin (CaM). The pCold vector system can also be used to selectively enrich target proteins with isotopes to study their properties in cell lysates using NMR spectroscopy. We have cloned 38 genes from a range of prokaryotic and eukaryotic organisms into both pCold and pET14 (ref. 3) systems, and found that pCold vectors are highly complementary to the widely used pET vectors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Schein, C.H. Solubility as a function of protein structure and solvent components. Biotechnology 8, 308–317 (1989).
Schirano, Y. & Shibata, D. Low temperature cultivation of Escherichia coli carrying a rice lipoxygenase L-2 cDNA produces a soluble and active enzyme at a high level. FEBS Lett. 271, 128–130 (1990).
Studier, F.W. & Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130 (1986).
Etchegaray, J.P. & Inouye, M. Translational enhancement by an element downstream of the initiation codon in Escherichia coli. J. Biol. Chem. 274, 10079–10085 (1999).
Mitta, M., Fang, L. & Inouye, M. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold-shock induction. Mol. Microbiol. 26, 321–335 (1997).
Xia, B., Etchegaray, J.P. & Inouye, M. Nonsense Mutations in cspA Cause Ribosome Trapping Leading to Complete Growth Inhibition and Cell Death at Low Temperature in Escherichia coli. J. Biol. Chem. 276, 35581–35588 (2001).
Bae, W., Jones, P.G. & Inouye, M. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J. Bacteriol. 179, 7081–7088 (1997).
Zhang, M., Tanaka, T. & Ikura, M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 2, 758–767 (1995).
Wishart, D.S. & Sykes, B.D. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4, 171–180 (1994).
Gronenborn, A.M. & Clore, G.M. Rapid screening for structural integrity of expressed proteins by heteronuclear NMR spectroscopy. Protein Sci. 5, 174–177 (1996).
Erbel, P.J. et al. Identification and biosynthesis of cyclic enterobacterial common antigen in Escherichia coli. J. Bacteriol. 185, 1995–2004 (2003).
Almeida, F.C. et al. Selectively labeling the heterologous protein in Escherichia coli for NMR studies: a strategy to speed up NMR spectroscopy. J. Magn. Reson. 148, 142–146 (2001).
Wunderlich, Z. et al. The protein target list of the Northeast Structural Genomics Consortium. (in the press) (2004). Published online May 2004.
Jansson, M. et al. High level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. Biomol. NMR 7, 131–141 (1996).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Tanaka, T. et al. NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ. Nature 396, 88–92 (1998).
Cavanaugh, J., Fairbrother, W.J., Palmer, A.G., III & Skelton, N.J. Protein NMR Spectroscopy (Academic Press, San Diego, 1996).
Montelione, G.T., Rios, C.B., Swapna, G.V.T. & Zimmerman, D.E. NMR pulse sequences and computational approaches for analysis of sequence-specific backbone resonance assignments of proteins. in Biological Magnetic Resonance (Kluwer Academic/Plenum, New York, 1999) 17, 81–130.
Acknowledgements
We thank Y. Chiang for helpful discussions. This work was supported by NIH grants R21-GM067061 (to M.I.) and P50-GM62413 (to G.T.M.), a grant from Takara-Bio Inc., Japan (to M.I.), and a Canadian Institutes of Health Research (CIHR) grant (to M.I.). M.I. is a CIHR senior investigator.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
SDS-PAGE patterns of FR genes expressed in pET14 and pColdI vectors (see Table 1) (PDF 140 kb)
Supplementary Table 1
Summary of triple-resonance NMR experiments and key parameters used for determining backbone resonance assignments for Xenopus calmodulin in whole-cell lysate. (PDF 7 kb)
Supplementary Table 2
Sequence specific backbone 1H, 15N, and 13C, and 13Cβ resonance assignments a for Xenopus calmodulin at pH 6.8 and temperature of 10 °C, determined by triple resonance NMR experiments on whole-cell lysates. (PDF 11 kb)
Rights and permissions
About this article
Cite this article
Qing, G., Ma, LC., Khorchid, A. et al. Cold-shock induced high-yield protein production in Escherichia coli. Nat Biotechnol 22, 877–882 (2004). https://doi.org/10.1038/nbt984
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt984
This article is cited by
-
Molecular Biology Applications of Psychrophilic Enzymes: Adaptations, Advantages, Expression, and Prospective
Applied Biochemistry and Biotechnology (2024)
-
Improving expression and assembly of difficult-to-express heterologous proteins in Saccharomyces cerevisiae by culturing at a sub-physiological temperature
Microbial Cell Factories (2023)
-
Omics-guided bacterial engineering of Escherichia coli ER2566 for recombinant protein expression
Applied Microbiology and Biotechnology (2023)
-
New temperature-switchable acyl homoserine lactone-regulated expression vector
Applied Microbiology and Biotechnology (2023)
-
A New Hybrid Protein Is a Novel Regulator for Experimental Colitis in Rats
Inflammation (2022)