Abstract
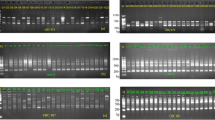
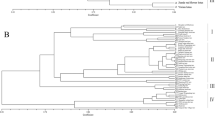
DNA oligonucleotide and amplification fingerprinting have been successfully used to detect genetic polymorphisms in 15 representative species and cultivars of the genus Musa, comprising AA, AAA, AAAA, AAB, ABB, and BB genotypes. In–gel–hybridization of Hinf I–digested genomic banana DNA to the 32P–labeled synthetic oligonucleotides (GATA)4, (GTG)5, and (CA)8 revealed considerable polymorphisms between Musa species and cultivars. The fingerprint patterns proved to be somatically stable and did not show differences between individual plants of “Grand Nain” (AAA genotype). Dendrograms based on oligonucleotide fingerprint band sharing data proved to be consistent with most of the known features of the history of banana and plantain cultivation and evolution, respectively. DNA samples from the same banana species and cultivars were also amplified by PCR using single or pairwise combinations of short oligonucleotide primers. Amplification products were separated on agarose or polyacrylamide gels and visualized by ethidium bromide or silver staining, respectively. Polymorphic patterns were obtained with some but not all primers. By using the CCCTCTGCGG primer in simplex and/or duplex PCR, the induced mutant ‘GN60A’ was clearly recognized from its original variety ‘Grand Nain’. Both fingerprint techniques allowed the detection of bands characteristic for the A and B genome. This DNA fingerprinting technology has potential application in several areas of Musa improvement.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Stover, P. and Simmonds, N.W. 1987. Bananas. Longman Sci. and Techn., Harlow, UK.
Simmonds, N.W. 1962. The Evolution of Bananas. Longman, London, UK.
Novak, F.J. 1992. Musa (Bananas and Plantains), p. 449–488. In: Biotechnology of Perennial Fruit Crops. Hammerschlag, F. A. and Litz, R. (Eds.), C.A.B. Intern., Wallington, Oxon.
Tautz, D. and Renz, M. 1984. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucl. Acids Res. 12: 4127–4138.
Weising, K., Ramser, J., Kaemmer, D., Kahl, G. and Epplen, J.T. 1991. Oligonucleotide fingerprinting in plants and fungi, p. 312–319. In: DNA Fingerprinting: Approaches and Applications. Burke, T., Dolf, G., Jeffreys, A. J. and Wolff, R. (Eds.). Birkher, Basel, Switzerland.
Epplen, J.T., Ammer, H., Epplen, C., Kammerbauer, C. et al. 1991. Oligonucleotide fingerprinting using simple repeat motifs: a convenient, ubiquitously applicable method to ddetect hypervariability for multiple purposes, p. 50–69. Ibid.
Caetano-Anollés, G., Bassam, B.J. and Gresshoff, P.M. 1991. DNA amplification fingerprinting using very short arbitrary Oligonucleotide primers. Bio/Technology 9: 553–557.
Williams, J.G.K., Kubelik, A.R., Litvak, K.J., Rafalski, J.A. and Tingey, S.V. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucl. Acids Res. 18: 6531–6535.
Welsh, J. and McClelland, M. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucl. Acids Res. 18: 7213–7218.
Weising, K., Fiala, B., Ramloch, K., Kahl, G. and Epplen, J.T. 1990. Oligonucleotide fingerprinting in angiosperms. Fingerprint News 2(2): 5–8.
Valmayor, R.V., Rivera, F.N. and Lomuljo, F.M. 1981. Philippine banana cultivar names and synonyms. Nat. Plant Genetic Res. Lab., Inst. Plant Breeding, Bull. No. 3, University Philippines, Los Banos.
Simmonds, N.W. and Weatherup, H. 1990. Numerical taxonomy of cultivated bananas. Trop. Agric. (Trinidad) 67: 90–92.
Weising, K., Beyermann, B., Ramser, J. and Kahl, G. 1991. Plant DNA fingerprinting with radioactive and digoxigenated Oligonucleotide probes complementary to simple repetitive DNA sequences. Electrophoresis 12: 159–169.
Weising, K., Kaemmer, D., Weigand, F., Epplen, J.T. and Kahl, G. 1992. Oligonucleotide fingerprinting reveals various probe-dependent levels of informativeness in chick-pea (Cicer arietinum). Genome 35, In Press.
Welsh, J. and McClelland, M. 1991. Genomic fingerprinting using arbitrarily primed PCR and a matrix of pairwise combinations of primers. Nucl. Acids Res. 19: 5275–5279.
Bell, D.A. and DeMarini, D.M. 1991. Excessive cycling converts PCR products to random-length higher molecular weight fragments. Nucl. Acids Res. 19: 5079.
Horn, G.T., Richards, B. and Klinger, K.W. 1989. Amplification of a highly polymorphic VNTR segment by the polymerase chain reaction. Nucl. Acids Res. 17: 2140.
Felsenstein, J. 1990. PHYLIP Manual Version 3.3. University of California, Berkeley, CA.
Schmid, J., Voss, E. and Soil, D.R. 1990. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J. Clin. Microbiol. 28: 1236–1243.
Kistler, H.C., Momol, E.A. and Benny, U. 1991. Repetitive genomic sequences for determining relatedness among strains of Fusarium oxy-sporum . Phytopathology 81: 331–336.
Van Heusden, A.W. and Bachmann, K. 1992. Genotype relationships in Microseris elegans (Asteraceae, Lactuceae) revealed by DNA amplification from arbitrary primers (RAPDs). Pl. Syst. Evol. 179: 221–233.
INIBAP/IBFGR. 1990. Musa conservation and documentation: Proceedings of a workshop held in Leuven, Belgium.
Stover, R.H. and Buddenhagen, I.W. 1986. Banana breeding: polyploidy, disease resistance and productivity. Fruits 41: 175–191.
Ali, S., Müller, C.R. and Epplen, J.T. 1986. DNA fingerprinting by Oligonucleotide probes specific for simple repeats. Hum. Genet. 74: 239–243.
Schäfer, R., Zischler, H., Birsner, U., Becker, A. and Epplen, J.T. 1988. Optimized probes for DNA fingerprinting. Electrophoresis 9: 369–374.
Thein, S.L. and Wallace, R.B. 1986. The use of synthetic oligonucleotides as specific hybridization probes in the diagnosis of genetic disorders, p. 33–50. In: Human Genetic Diseases, A Practical Approach. Davies, K. (Ed.). IRL Press, Oxford, UK.
Novak, F.J., Afza, R., van Duren, M. and Omar, M.S. 1990. Mutation induction by gamma irradiation of in vitro cultured shoot tips of banana and plantain (Musa cvs). Trop. Agric. (Trinidad) 67: 21–28.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Kaemmer, D., Afza, R., Weising, K. et al. Oligonucleotide and Amplification Fingerprinting of Wild Species and Cultivars of Banana (Musa spp.). Nat Biotechnol 10, 1030–1035 (1992). https://doi.org/10.1038/nbt0992-1030
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0992-1030