Abstract
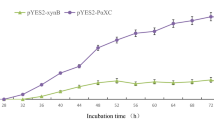
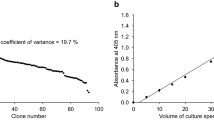
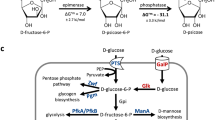
Mouse pancreatic α–amylase complementary DNA was inserted into a yeast shuttle vector after the Saccharomyces cerevisiae MFα1 promoter and secretion signals coding sequences. When transformed with the recombinant plasmid, S. cerevisiae cells were able to synthesize and secrete functional α–amylase, efficiently hydrolysing starch present in the culture medium. Stable amylolytic cells were obtained from different yeast strains. This work represents a significant step towards producing yeast that can convert starchy materials directly to ethanol.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Thomsen, K.K. 1983. Mouse α-amylase synthesized by Saccharomyces cerevisiae is released into the culture medium. Carlsberg Res. Commun. 48:545–555.
Rothstein, S.J., Lazarus, C.M., Smith, W.E., Baulcombe, D.C., and Gatenby, A.A. 1984. Secretion of a wheat α-amylase expressed in yeast. Nature 308:662–665.
Sandstedt, R.M. and Ueda, S. 1969. Alpha-amylase adsorption on raw starch and its relation to raw starch digestion. J. Japan Soc. Starch Sci. 17:215–228.
Ueda, S. 1982. Raw starch digestion and ethanol fermentation of starch materials without cooking by fungal amylases. Workshop on Carbohydrate Resources and Biotechnology, Tsukuba, Japan, p. 16–24.
Aunstrup, K. 1978. Enzymes of industrial interest: Traditional products. Annual Rep. on Ferment. Processes 2:125–154.
Tosi, M., Bovey, R., Astolfi Filho, S., Bodary, S., Meisler, M., and Wellauer, P. 1984. Multiple non-allelic genes encoding pancreatic α-amylase of mouse are expressed in a strain-specific fashion. EMBO J. 3:2809–2816.
Cornelis, P., Digneffe, C., and Willemot, K. 1982. Cloning and expression of a Bacillus coagulans amylase gene in Escherichia coli. Molec. Gen. Genet. 186:507–511.
Astolfi Filho, S., Tosi, M., Azevedo, M.O., Galembeck, E.V., and Schenberg Frascino, A.C. 1983. Construção de um clone de Escherichia coli capaz de sintetizar α-amylase de camundongo. Arq. Biol. Tecnol. 26:151.
Novick, P., Field, C., and Schekman, R. 1980. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21:205–215.
Duntze, W., MacKay, V., and Manney, T.R. 1970. Saccharomyces cerevisiae: A diffusible sex factor. Science 168:1472–1473.
Kurjan, J. and Herskowitz, I. 1982. Structure of a yeast pheromone gene (MFα): A putative α-factor precursor contains four tandem copies of mature α-factor. Cell 30:933–943.
Singh, A., Chen, E.Y., Lugovoy, J.M., Chang, C.N., Hitzeman, R.A., and Seeburg, P.H. 1983. Saccharomyces cerevisiae contains two discrete genes coding for the α-factor pheromone. Nucleic Acids Res. 11:4049–4063.
Julius, D., Blair, L., Brake, A., Sprague, G., and Thorner, J. 1983. Yeast α-factor is processed from a larger precursor polypeptide: The essential role of a membrane-bound dipeptidyl aminopeptidase. Cell 32:839–852.
Julius, D., Schekman, R., and Thorner, J. 1984. Glycosylation and processing of prepro-α-factor through the yeast secretory pathway. Cell 36:309–318.
Bitter, G.A., Chen, K.K., Banks, A.R., and Lai, P.H. 1984. Secretion of foreign proteins from Saccharomyces cerevisiae directed by α-factor gene fusions. Proc. Natl. Acad. Sci. (USA) 81:5530–5534.
Brake, A.J., Merryweather, J.P., Coit, D.G., Heberlein, U.A., Masiarz, F.R., Mullenbach, G.T., Urdea, M.S., Valenzuela, P., and Barr, P.J. 1984. α-factor-directed synthesis and secretion of mature foreign proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. (USA) 81:4642–4646.
Singh, A., Lugovoy, J.M., Kohr, W.J. and Perry, L.J. 1984. Synthesis, secretion and processing of α-factor-interferon fusion proteins in yeast. Nucleic Acids Res. 12:8927–8938.
Broach, J.R., Strathern, J.N., and Hicks, J.B. 1979. Transformation in yeast: Development of a hybrid cloning vector and isolation of the CAN1 gene. Gene 8:121–133.
Zaret, K.S. and Sherman, F. 1982. DNA sequence required for efficient transcription termination in yeast. Cell 28:563–573.
Hitzeman, R.A., Leung, D.W., Perry, L.J., Kohr, W.J., Levine, H.L., and Goeddel, D.V. 1983. Secretion of human interferons by yeast. Science 219:620–625.
Falco, S.C., Rose, M., and Botstein, D. 1983. Homologous recombination between episomal plasmids and chromosomes in yeast. Genetics 105:843–856.
Innis, M.A., Holland, M.J., McCabe, P.C., Cole, G.E., Wittman, V.P., Tal, R., Watt, K.W.K., Gelfand, D.H., Holland, J.P., and Meade, J.H. 1985. Expression, glycosylation, and secretion of an Aspergillus glucoamylase by Saccharomyces cerevisiae. Science 228:21–26.
Wilson, J.J. and Ingledew, W.M. 1982. Isolation and characterization of Schwanniomyces alluvius amylolytic enzymes. Appl. Environ. Microbiol. 44:301–307.
Bolivar, F. and Bachman, K. 1979. Plasmids of Escherichia coli as cloning vectors. Meth. Enzym. 68:245–267.
Miller, J. 1972. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, New York.
Sherman, F., Fink, G.R., and Hicks, J.B. 1981. In: Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
Birnboim, H.C. and Doly, J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513–1523.
Maniatis, T., Fritsch, E., and Sambrook, J. 1982. Molecular Cloning, a Laboratory Manual. Cold Spring Harbor Laboratory, New York.
Sanger, F., Nicklen, S., and Coulson, A.R. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. (USA) 74:5463–6467.
Kilmartin, J.V., Wright, B., and Milstein, C. 1982. Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol. 93:576–582.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Filho, S., Galembeck, E., Faria, J. et al. Stable Yeast Transformants that Secrete Functional α–Amylase Encoded by Cloned Mouse Pancreatic cDNA. Nat Biotechnol 4, 311–315 (1986). https://doi.org/10.1038/nbt0486-311
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0486-311
This article is cited by
-
Extraction, Purification and Characterization of Thermostable, Alkaline Tolerant α-Amylase from Bacillus cereus
Indian Journal of Microbiology (2011)