Abstract
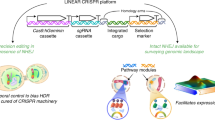
Our understanding of how genotype controls phenotype is limited by the scale at which we can precisely alter the genome and assess the phenotypic consequences of each perturbation. Here we describe a CRISPR–Cas9-based method for multiplexed accurate genome editing with short, trackable, integrated cellular barcodes (MAGESTIC) in Saccharomyces cerevisiae. MAGESTIC uses array-synthesized guide–donor oligos for plasmid-based high-throughput editing and features genomic barcode integration to prevent plasmid barcode loss and to enable robust phenotyping. We demonstrate that editing efficiency can be increased more than fivefold by recruiting donor DNA to the site of breaks using the LexA–Fkh1p fusion protein. We performed saturation editing of the essential gene SEC14 and identified amino acids critical for chemical inhibition of lipid signaling. We also constructed thousands of natural genetic variants, characterized guide mismatch tolerance at the genome scale, and ascertained that cryptic Pol III termination elements substantially reduce guide efficacy. MAGESTIC will be broadly useful to uncover the genetic basis of phenotypes in yeast.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
European Nucleotide Archive
Referenced accessions
Protein Data Bank
References
Garst, A.D. et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 35, 48–55 (2017).
Sadhu, M.J. et al. Highly parallel genome variant engineering with CRISPR/Cas9 in eukaryotic cells. Preprint at https://www.biorxiv.org/search/147637 (2017).
Guo, X. et al. High-throughput creation and functional profiling of eukaryotic DNA sequence variant libraries using CRISPR/Cas9. Preprint at https://www.biorxiv.org/search/195776 (2017).
Michlits, G. et al. CRISPR-UMI: single-cell lineage tracing of pooled CRISPR-Cas9 screens. Nat. Methods 14, 1191–1197 (2017).
DiCarlo, J.E. et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343 (2013).
Ugolini, S. & Bruschi, C.V. The red/white colony color assay in the yeast Saccharomyces cerevisiae: epistatic growth advantage of white ade8-18, ade2 cells over red ade2 cells. Curr. Genet. 30, 485–492 (1996).
Doench, J.G. et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014).
Doench, J.G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Clikeman, J.A., Khalsa, G.J., Barton, S.L. & Nickoloff, J.A. Homologous recombinational repair of double-strand breaks in yeast is enhanced by MAT heterozygosity through yKU-dependent and -independent mechanisms. Genetics 157, 579–589 (2001).
Wu, X. & Haber, J.E.A. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell 87, 277–285 (1996).
Sun, K., Coïc, E., Zhou, Z., Durrens, P. & Haber, J.E. Saccharomyces forkhead protein Fkh1 regulates donor preference during mating-type switching through the recombination enhancer. Genes Dev. 16, 2085–2096 (2002).
Dummer, A.M. et al. Binding of the Fkh1 Forkhead Associated Domain to a phosphopeptide within the Mph1 DNA helicase regulates mating-type switching in budding yeast. PLOS Genet. 12, e1006094 (2016).
Li, J. et al. Regulation of budding yeast mating-type switching donor preference by the FHA domain of Fkh1. PLoS Genet. 8, e1002630 (2012).
Chayakulkeeree, M. et al. SEC14 is a specific requirement for secretion of phospholipase B1 and pathogenicity of Cryptococcus neoformans. Mol. Microbiol. 80, 1088–1101 (2011).
Nile, A.H. et al. PITPs as targets for selectively interfering with phosphoinositide signaling in cells. Nat. Chem. Biol. 10, 76–84 (2014).
Fang, M. et al. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 15, 6447–6459 (1996).
Li, X. et al. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 157, 63–77 (2002).
Cleves, A.E. et al. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell 64, 789–800 (1991).
Fu, B.X.H., St Onge, R.P., Fire, A.Z. & Smith, J.D. Distinct patterns of Cas9 mismatch tolerance in vitro and in vivo. Nucleic Acids Res. 44, 5365–5377 (2016).
Hsu, P.D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Arimbasseri, A.G., Rijal, K. & Maraia, R.J. Transcription termination by the eukaryotic RNA polymerase III. Biochim. Biophys. Acta 1829, 318–330 (2013).
Braglia, P., Percudani, R. & Dieci, G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 280, 19551–19562 (2005).
Orioli, A. et al. Widespread occurrence of non-canonical transcription termination by human RNA polymerase III. Nucleic Acids Res. 39, 5499–5512 (2011).
Smith, J.D. et al. A method for high-throughput production of sequence-verified DNA libraries and strain collections. Mol. Syst. Biol. 13, 913 (2017).
Savic, N. et al. Covalent linkage of the DNA repair template to the CRISPR/Cas9 complex enhances homology-directed repair. Preprint at https://www.biorxiv.org/search/218149 (2017).
Ma, M. et al. Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res. 27, 578–581 (2017).
Gu, B., Posfai, E. & Rossant, J. Efficient generation of targeted large insertions in mouse embryos using 2C-HR-CRISPR. Preprint at https://www.biorxiv.org/search/204339 (2017).
Polo, S.E. & Jackson, S.P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25, 409–433 (2011).
Chen, J.S. et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410 (2017).
Kleinstiver, B.P. et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Slaymaker, I.M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Hu, J.H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
Ryan, O.W. et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife 3, e03703 (2014).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Gietz, R.D. & Schiestl, R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 (2007).
Xing, X. & Wei, W. SICtools: Find SNV/Indel differences between two bam files with near relationship. R package version 1.8.0. (2014). http://bioconductor.org/packages/SICtools/.
Sha, B., Phillips, S.E., Bankaitis, V.A. & Luo, M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature 391, 506–510 (1998).
Jones, G., Willett, P., Glen, R.C., Leach, A.R. & Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748 (1997).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2018).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D.J., Chen, Y. & Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Wickham,, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Waskom, M. et al. seaborn: v0.7.1 (June 2016) (Zenodo, 2016). doi:10.5281/zenodo.54844.
Acknowledgements
This work was supported by grants from the US National Institutes of Health (P01HG000205 to L.M.S. and R.W.D., R01GM121932-01A1 to R.P.S., U01GM110706-02 to R.W.D., RO1GM61766 to J.E.H., and RO1GM44530 to V.A.B.), the National Institute of Standards and Technology (70NANB15H268 to M.L.S.), and the European Research Council Advanced Investigator Grant (AdG-294542 to L.M.S.). K.R.R. was supported by a National Research Council postdoctoral fellowship. A.T. and V.A.B. were supported by the Robert A. Welch Foundation (award BE-0017). S.C.V. was supported by a Swiss National Science Foundation postdoctoral fellowship (P2EZP3_165220). Certain commercial equipment, instruments, or materials are identified in this document. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the products identified are necessarily the best available for the purpose. We thank the EMBL Genomics Core Facility for support and optimization of barcode sequencing protocols. This work is dedicated to the memory of Joe Horecka (12/1/1963-10/20/2017).
Author information
Authors and Affiliations
Contributions
K.R.R., J.D.S., S.C.V., R.P.S., and L.M.S. conceived and designed the study, and wrote and edited the paper. K.R.R., J.D.S., S.C.V., and R.P.S. performed experiments and analyzed data. K.R.R., S.C.V., G.L., and A.R.L. analyzed NGS data; C.S.T., A.C., S.S., M.N., J.H., W.T.B., M.A.M., J.S., and K.M.O. performed experiments. A.T. and V.A.B. performed computational structural analysis on Sec14p-NPPM; W.W. performed variant calling for the different yeast strains. J.E.H. suggested adapting the LexA–Fkh1p system to the guide–donor plasmid. R.S.A., R.W.D., and M.L.S. advised the study. R.P.S. and L.M.S. were responsible for the coordination of the study. All authors read, corrected, and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
K.R.R., J.D.S., J.E.H., R.P.S. and L.M.S. have filed a provisional application (US 62/559,493) with the US Patent and Trademark Office on this work.
Integrated supplementary information
Supplementary Figure 1 Barcode and feature representation throughout library construction
(a) Venn diagram representation of barcode (left) and feature (right) overlap among the oligo pool, step 1, and step 2 libraries. Note that barcodes are added during PCR amplification of the initial oligo library and therefore cannot be analyzed until cloning of step 1 libraries. Although most barcodes observed in step 1 libraries are not recovered in step 2 or yeast libraries, 88,821 out of 100,000 designed features are observed in the yeast pre-editing library (see Methods). (b) Representation of barcodes (left) and features (right) at the indicated stages of library construction as a function of subsampling reads from 0 to 20 million (left) or 10 million (right) reads. The barcodes/features are plotted as a percentage relative to all barcodes/features identified in their respective initial reference pools (step 1 cloning for barcodes and oligo pool for features). (c) Representation of barcodes (left) and features (right) at the indicated stages of library construction as a function of what percentage of the reads (Y) in each sample are attributed to what percentage (X) of the top barcodes/features in each sample, where the barcodes/features are sorted from highest to lowest abundance from left to right on the x-axis. The dotted line (slope = 1) depicts an idealized library of perfect uniformity where all members are present at equal abundance.
Supplementary Figure 2 Guide X promoter comparison and integration kinetics
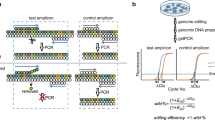
Three different promoters were tested to drive the expression of guide X: RPR1, SNR52, and the tRNA (Tyr)-HDV ribozyme promoter. As in Fig. 2b, integration of the guide-donor barcode at each generation was assayed by amplification with primers flanking the chromosomal barcode locus. The larger amplicon size indicates successful integration of the guide-donor barcode. The integration kinetics were tested for the single ADE2 guide-donor plasmid. The self-destruction of the guide-donor plasmids was assessed by a three-primer PCR, with a common forward primer and either a guide-donor plasmid-specific primer (top band) or a Cas9-plasmid specific primer (bottom band). The experiment was conducted with biological replicates from independent guide-donor transformations with similar kinetics of barcode integration.
Supplementary Figure 3 Detailed synonymous codon spreading strategy to enable mutation of codons outside of guide RNA recognition regions
Amino acid saturation editing strategy for open reading frames with (a) target codons that fall within the 20 bp of NGG-PAM guide recognition sequences and (b) target codons that fall outside of the nearest NGG-PAM guide recognition sequence. The examples show the design of the donor DNA to accompany guide RNA utilizing the nearest downstream PAM. The nonsynonymous changes (red) are accompanied by synonymous changes (cyan) that spread towards the Cas9 cleavage site (3 bp upstream of the PAM). The synonymous changes block cleavage of the donor by the Cas9::guide complex and minimize microhomology between the nonsynonymous variant of interest and the guide-disrupting changes. A pseudo-WT control is included to rule out effects due to the synonymous changes.
Supplementary Figure 4 Fitness effects of all possible amino acid mutations across a region of Sec14p
(a) The relative abundance of the indicated nonsynonymous changes from amino acid positions 102 to 137 in Sec14p after editing in the haploid suppressor and mating to the complementary suppressor (see Fig. 4b; * = stop codon). (b) Log10 values of the normalized read counts for each of 1361 variants observed in the haploid suppressor background (in which SEC14 is non-essential; x=axis) vs. the diploid (in which SEC14 is essential by virtue of suppressor complementation; y-axis). Variants encoding premature termination codons (PTCs) are represented by green points. The Pearson’s correlation coefficient was obtained using the Python scipy.stats.pearsonr (version 1.0.0) function with default parameters and a 2-tailed p-value.
Supplementary Figure 5 Drug fitness replicate correlations with upstream and downstream synonymous codon changes for each SEC14 variant
(a) Log10 values of the normalized read counts are plotted for two replicate samples after 12 generations of growth in the presence of NPPM (n = 969 variants with read counts > 0 in at least one replicate). (b) The log2-fold change for each variant in the presence of NPPMs relative to the DMSO-control is plotted for the upstream synonymous changes version of each variant (y-axis) vs. the downstream synonymous changes version (x-axis). The Pearson correlation coefficient is indicated by pearsonr as in Supplementary Fig. 4b (n = 797 variants, filtered for amino acid variants with at least 10 reads for both upstream and downstream synonymous versions).
Supplementary Figure 6 Azimuth score and PAM identity are correlated with guide efficacy
(a) The Azimuth score for each guide in our dataset (x-axis) is plotted against log2-fold change after editing (y-axis) (Spearman rho -0.18, Pearson R = -0.19, both p < 2.2E-16). The trend line derives from a linear regression of logFC to Azimuth score (R2 = 0.037, p<2.2E-16). The color intensity depicts the count of barcodes per box (N = 23,866 barcodes). (b) Violin plots of the effect of PAM on log2-fold change after editing (black dots depict medians and black lines indicate the 25th and 75th percentiles). The number of barcodes (N) for each PAM type is indicated in the above plots. A one-sided Wilcoxon test was used for between-group comparisons; for each group, location shift and 99% confidence intervals (in square brackets) are as follows: TGG_AGG: 0.12 [0.05, Inf], p = 6.478E-05; TGG_CGG: 0.19 [0.1, Inf], p = 7.72E-07; TGG_GGG: 0.31 [0.22, Inf], p = 6.744E-16. Dead guides are shown for comparison.
Supplementary Figure 7 Effect of homopolymers and T-homopolymer location on guide RNA efficacy
(a) Log2-fold changes for guide-donor barcodes post-editing relative to pre-editing is plotted as a function of the homopolymer content. For each guide, the longest homopolymer for each of the four nucleotides was identified, and guide-donor barcodes were grouped into the bins of the designated length. Dead guides are shown for comparison. Box plot boundaries depict the upper and lower quartile, with the line denoting the median value. The number of barcodes (N) for each group is specified above the boxes. (b) The log2-fold change for TTT and TTTT-containing guides post-editing relative to pre-editing is plotted as a function of whether the T-stretch is at the 3’ end of the guide (*** p = 1.97E-05, * p = 0.01213). Note that the Cas9 guide RNA scaffold used in this study begins with GTTTA. A one-sided Wilcoxon test was used for between-group comparisons; for each group, location shift and 99% confidence intervals (in square brackets) are as follows: T3: 0.36 [0.16, Inf], T4: 0.26 [-0.01, Inf]. Dead guides and guides containing T-homopolymers less than 3 nucleotides long are plotted for comparison. The number of barcodes in each group is specified above the boxes.
Supplementary Figure 8 Relationship between Azimuth score, T-score, and guide efficacy
(a) The Azimuth score for each guide in our dataset is plotted against log2-fold change in barcode abundance after editing. Each barcode is colored according to the T-score of its associated guide. Grey dots denote efficient guides (T-score < 5, N = 20,474), salmon dots indicate guides with T5 or imperfect T-stretches (T-score 5 – 6.5, N = 2787) showing slight enrichment during editing and red dots denote very inefficient guides (T-score >=7, N = 332) showing more pronounced enrichment during editing. (b) The Azimuth score for each guide in the Azimuth training set is plotted against the experimentally-determined guide efficacy. Each guide is colored according to T-score, with guides with T-score < 5 in grey (N = 5243) and guides with T-score 5 - 6.5 in salmon (N = 67). 6.5 was the highest T-score we observed in this dataset.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 (PDF 1354 kb)
Supplementary Tables
Supplementary tables 1, 3–4 (PDF 272 kb)
Supplementary Table 2
Growth analysis of individual Sec14p variants (XLSX 93 kb)
Rights and permissions
About this article
Cite this article
Roy, K., Smith, J., Vonesch, S. et al. Multiplexed precision genome editing with trackable genomic barcodes in yeast. Nat Biotechnol 36, 512–520 (2018). https://doi.org/10.1038/nbt.4137
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4137
This article is cited by
-
From beer to breadboards: yeast as a force for biological innovation
Genome Biology (2024)
-
Deep mutational scanning of essential bacterial proteins can guide antibiotic development
Nature Communications (2023)
-
A Cas3-base editing tool for targetable in vivo mutagenesis
Nature Communications (2023)
-
From systems to structure — using genetic data to model protein structures
Nature Reviews Genetics (2022)
-
Towards next-generation cell factories by rational genome-scale engineering
Nature Catalysis (2022)