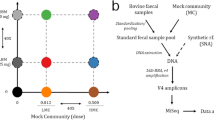
Abstract
Amplicon-based marker gene surveys form the basis of most microbiome and other microbial community studies. Such PCR-based methods have multiple steps, each of which is susceptible to error and bias. Variance in results has also arisen through the use of multiple methods of next-generation sequencing (NGS) amplicon library preparation. Here we formally characterized errors and biases by comparing different methods of amplicon-based NGS library preparation. Using mock community standards, we analyzed the amplification process to reveal insights into sources of experimental error and bias in amplicon-based microbial community and microbiome experiments. We present a method that improves on the current best practices and enables the detection of taxonomic groups that often go undetected with existing methods.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Cho, I. & Blaser, M.J. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012).
Gilbert, J.A., Jansson, J.K. & Knight, R. The Earth Microbiome project: successes and aspirations. BMC Biol. 12, 69 (2014).
The Human Microbiome Project Consortium A framework for human microbiome research. Nature 486, 215–221 (2012).
Jumpstart Consortium Human Microbiome Project Data Generation Working Group Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One 7, e39315 (2012).
Goodrich, J.K. et al. Conducting a microbiome study. Cell 158, 250–262 (2014).
Kuczynski, J. et al. Experimental and analytical tools for studying the human microbiome. Nat. Rev. Genet. 13, 47–58 (2012).
Caporaso, J.G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Schloss, P.D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Salter, S.J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
Brooks, J.P. et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 15, 66 (2015).
Pinto, A.J. & Raskin, L. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS One 7, e43093 (2012).
Sinha, R., Abnet, C.C., White, O., Knight, R. & Huttenhower, C. The microbiome quality control project: baseline study design and future directions. Genome Biol. 16, 276 (2015).
Zhou, J. et al. Random sampling process leads to overestimation of β-diversity of microbial communities. MBio 4, e00324–13 (2013).
Yuan, S., Cohen, D.B., Ravel, J., Abdo, Z. & Forney, L.J. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One 7, e33865 (2012).
Kennedy, N.A. et al. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One 9, e88982 (2014).
Feinstein, L.M., Sul, W.J. & Blackwood, C.B. Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl. Environ. Microbiol. 75, 5428–5433 (2009).
Zhao, J. et al. Effect of sample storage conditions on culture-independent bacterial community measures in cystic fibrosis sputum specimens. J. Clin. Microbiol. 49, 3717–3718 (2011).
Cardona, S. et al. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 12, 158 (2012).
Aird, D. et al. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12, R18 (2011).
Ahn, J.-H., Kim, B.-Y., Song, J. & Weon, H.-Y. Effects of PCR cycle number and DNA polymerase type on the 16S rRNA gene pyrosequencing analysis of bacterial communities. J. Microbiol. 50, 1071–1074 (2012).
Wu, J.-Y. et al. Effects of polymerase, template dilution and cycle number on PCR based 16 S rRNA diversity analysis using the deep sequencing method. BMC Microbiol. 10, 255 (2010).
Ishii, K. & Fukui, M. Optimization of annealing temperature to reduce bias caused by a primer mismatch in multitemplate PCR. Appl. Environ. Microbiol. 67, 3753–3755 (2001).
D'Amore, R. et al. A comprehensive benchmarking study of protocols and sequencing platforms for 16S rRNA community profiling. BMC Genomics 17, 55 (2016).
Kennedy, K., Hall, M.W., Lynch, M.D.J., Moreno-Hagelsieb, G. & Neufeld, J.D. Evaluating bias of Illumina-based bacterial 16S rRNA gene profiles. Appl. Environ. Microbiol. 80, 5717–5722 (2014).
Hansen, M.C., Tolker-Nielsen, T., Givskov, M. & Molin, S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol. Ecol. 26, 141–149 (1998).
Reysenbach, A.L., Giver, L.J., Wickham, G.S. & Pace, N.R. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58, 3417–3418 (1992).
Mao, D.-P., Zhou, Q., Chen, C.-Y. & Quan, Z.-X. Coverage evaluation of universal bacterial primers using the metagenomic datasets. BMC Microbiol. 12, 66 (2012).
Polz, M.F. & Cavanaugh, C.M. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64, 3724–3730 (1998).
Hong, S., Bunge, J., Leslin, C., Jeon, S. & Epstein, S.S. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 3, 1365–1373 (2009).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1 (2013).
Kozich, J.J., Westcott, S.L., Baxter, N.T., Highlander, S.K. & Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013).
Quail, M.A. et al. Optimal enzymes for amplifying sequencing libraries. Nat. Methods 9, 10–11 (2012).
Schloss, P.D., Gevers, D. & Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6, e27310 (2011).
Patin, N.V., Kunin, V., Lidström, U. & Ashby, M.N. Effects of OTU clustering and PCR artifacts on microbial diversity estimates. Microb. Ecol. 65, 709–719 (2013).
Haas, B.J. et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504 (2011).
Wagner, A. et al. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Syst. Biol. 43, 250–261 (1994).
Suzuki, M.T. & Giovannoni, S.J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62, 625–630 (1996).
Schirmer, M. et al. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 43, e37 (2015).
Zhou, H.-W. et al. BIPES, a cost-effective high-throughput method for assessing microbial diversity. ISME J. 5, 741–749 (2011).
Degnan, P.H. & Ochman, H. Illumina-based analysis of microbial community diversity. ISME J. 6, 183–194 (2012).
Gloor, G.B. et al. Microbiome profiling by illumina sequencing of combinatorial sequence-tagged PCR products. PLoS One 5, e15406 (2010).
Claesson, M.J. et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 38, e200 (2010).
Caporaso, J.G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012).
Fadrosh, D.W. et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2, 6 (2014).
Bartram, A.K., Lynch, M.D.J., Stearns, J.C., Moreno-Hagelsieb, G. & Neufeld, J.D. Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl. Environ. Microbiol. 77, 3846–3852 (2011).
Salipante, S.J. et al. Performance comparison of Illumina and ion torrent next-generation sequencing platforms for 16S rRNA-based bacterial community profiling. Appl. Environ. Microbiol. 80, 7583–7591 (2014).
Illumina 16S metagenomic sequencing library preparation (Illumina Technical Note 15044223 Rev. A). Illumina http://support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (2013).
Faith, J.J. et al. The long-term stability of the human gut microbiota. Science 341, 1237439 (2013).
Lundberg, D.S., Yourstone, S., Mieczkowski, P., Jones, C.D. & Dangl, J.L. Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10, 999–1002 (2013).
Lee, C.K. et al. Groundtruthing next-gen sequencing for microbial ecology-biases and errors in community structure estimates from PCR amplicon pyrosequencing. PLoS One 7, e44224 (2012).
Nelson, M.C., Morrison, H.G., Benjamino, J., Grim, S.L. & Graf, J. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PLoS One 9, e94249 (2014).
Brown, C.T. et al. Unusual biology across a group comprising more than 15% of domain bacteria. Nature 523, 208–211 (2015).
Eloe-Fadrosh, E.A., Ivanova, N.N., Woyke, T. & Kyrpides, N.C. Metagenomics uncovers gaps in amplicon-based detection of microbial diversity. Nat. Microbiol. 1, 15032 (2016).
Wang, G.C. & Wang, Y. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63, 4645–4650 (1997).
Wang, G.C. & Wang, Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 142, 1107–1114 (1996).
Lahr, D.J.G. & Katz, L.A. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. Biotechniques 47, 857–866 (2009).
Kunkel, T.A. & Bebenek, K. DNA replication fidelity. Annu. Rev. Biochem. 69, 497–529 (2000).
Ayyadevara, S., Thaden, J.J. & Shmookler Reis, R.J. Discrimination of primer 3′-nucleotide mismatch by taq DNA polymerase during polymerase chain reaction. Anal. Biochem. 284, 11–18 (2000).
Bru, D., Martin-Laurent, F. & Philippot, L. Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl. Environ. Microbiol. 74, 1660–1663 (2008).
Jones, M.B. et al. Library preparation methodology can influence genomic and functional predictions in human microbiome research. Proc. Natl. Acad. Sci. USA 112, 14024–14029 (2015).
Yu, Z. & Morrison, M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36, 808–812 (2004).
Bolger, A.M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Masella, A.P., Bartram, A.K., Truszkowski, J.M., Brown, D.G. & Neufeld, J.D. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13, 31 (2012).
Cock, P.J.A. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Crooks, G.E., Hon, G., Chandonia, J.-M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Acknowledgements
We thank the staff of the University of Minnesota Genomics Center for helpful discussions and technical support. This work was supported by the Minnesota Partnership for Biotechnology and Medical Genomics (grant MNP IF #14.09). This work was carried out in part using computing resources at the University of Minnesota Supercomputing Institute. This work was also supported by the Margot Marsh Biodiversity Foundation and the US National Institutes of Health (PharmacoNeuroImmunology Fellowship NIH/NIDA T32 DA007097-32 to J.B.C.).
Author information
Authors and Affiliations
Contributions
D.M.G. and K.B.B. conceived and designed the experiments, analyzed data, and wrote the manuscript; J.G. and T.J.G. contributed to the analysis; P.V. and D.K. carried out the modeling and helped write the manuscript; D.M.G., A.M., A.H., and A.B. conducted the experiments; J.B.C., T.J.J., and R.H. contributed experimental samples.
Corresponding author
Ethics declarations
Competing interests
D.M.G. and K.B.B. are inventors on a provisional patent application filed with the USPTO (62/332,879) that incorporates aspects of the findings described here.
Integrated supplementary information
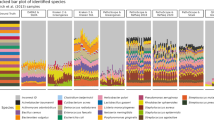
Supplementary Figure 1 The effect of method and enzyme choice on the accuracy of 16S rRNA gene microbiome profiling.
A-G) Bar plots showing observed even mock community mean abundances (HM-276D, unless otherwise stated) measured using the following methods (Expected abundances are indicated with the dashed line. Black asterisks indicate that the observed abundance deviated by more than 5-fold from the expected value. Red asterisks indicate taxa that had no mapped reads (drop-outs). Error bars are +/- SEM):
A) Reported by Kozich et al.,1 n = 12. § Mapped to the HM-278D reference file.
B) The EMP protocol, reported by Nelson et al.,2 n = 2.
C) The EMP protocol (this study), n = 3.
D) The EMP protocol, substituting KAPA HiFi polymerase for the standard Taq polymerase, n = 3.
E) The Dual-indexing (DI) protocol with Taq polymerase, n = 4.
F) The DI protocol with Q5 polymerase, n = 4.
G) The DI protocol with KAPA HiFi polymerase, n = 4.
H) Mean Absolute Percentage Error (MAPE) plot for the HM-276D even mock community data measured using the indicated methods. § HM-278D expected abundance values were used to calculate MAPE for this data set. Error bars are +/- SEM.
I) Scatter plot comparing HM-276D even mock community data reported by Nelson et al.2 using the EMP protocol to data collected for this study using the EMP protocol. Error bars are +/- SEM.
J) Average number of L6 (genus level) taxa observed with the indicated methods. Error bars are +/- SEM. *** p < 0.01 determined by ANOVA with Tukey HSD post-hoc test.
K-O) Bar plots showing observed HM-277D staggered mock mean abundances versus expected abundances measured using the following methods (Expected abundances are indicated with the dashed line. Black asterisks indicate that the observed abundance deviated by more than 5-fold from the expected value. Red asterisks indicate taxa that had no mapped reads (drop-outs). Star indicates error bar with a lower bound of zero that cannot be plotted on a log scale. Error bars are +/- SEM):
K) The EMP protocol, n = 3.
L) The EMP protocol, substituting KAPA HiFi polymerase for the standard Taq polymerase, n = 3.
M) The Dual-indexing (DI) protocol with Taq polymerase, n = 3.
N) The DI protocol with Q5 polymerase, n = 3.
O) The DI protocol with KAPA HiFi polymerase, n = 3.
P) MAPE plot for the HM-277D staggered mock community data measured using the indicated methods. Error bars are +/- SEM.
Supplementary Figure 2 The effect of annealing temperature on accuracy, chimera formation, and sample balance.
Plots for the HM-276D even mock community at 5 different starting template concentrations amplified for 35 cycles at either 50°C or 55°C using KAPA HiFi, Q5, and Taq polymerase showing:
A-C) Root mean square deviation (RMSD).
D-F) Percentage of chimeric reads.
G-I) Total number of reads.
Supplementary Figure 3 The effect of KAPA HiFi enzyme concentration on accuracy, chimera formation, sample balance, and adaptor dimer formation.
Plots for the HM-276D even mock community at 5 different starting template concentrations amplified for 20, 25, 30, or 35 cycles using 0.25x, 0.5x, 1x KAPA HiFi polymerase, or KAPA ReadyMix showing:
A-D) Root mean square deviation (RMSD).
E-H) Percentage of chimeric reads.
I-J) Total number of reads.
M-P) Percentage of adapter dimers.
Supplementary Figure 4 Primer editing artifacts.
A) Distribution of edited bases in the V4 515F primer region in data from a pure isolate of Campylobacter jejuni measured with the DI protocol with KAPA ReadyMix.
B) Distribution of edited bases in the V4 806R primer region in data from a pure isolate of Campylobacter jejuni measured with the DI protocol with KAPA ReadyMix.
C) Schematic of 16S V3-V5 amplification from a pure isolate of Campylobacter jejuni. This amplicon contains the V4 515F primer sequence, allowing assessment of the endogenous sequence.
D) Percentage of each base observed at position 6 of the sequence corresponding to the V4 515F primer sequence in a V3-V5 amplicon from a pure isolate of Campylobacter jejuni.
Supplementary Figure 5 Recovery of an organism with primer mismatches depends both on the use of a proofreading polymerase and on the use of sequencing primers that do not overlap with the initial amplification primers.
A) When using standard Taq polymerase and custom sequencing primers, organisms with a critical mismatch to the amplification primers are neither expected to be amplified in the enrichment PCR, nor targeted by the custom sequencing primer in the sequencing reaction. Right column, percentage of P. acnes observed using such amplification and sequencing conditions.
B) With a proofreading polymerase and custom sequencing primers, organisms with a critical mismatch to the amplification primers are amplified in the enrichment PCR, but such amplicons have a mismatch to the custom sequencing primer in the sequencing reaction. Right column, percentage of P. acnes observed using such amplification and sequencing conditions.
C) Since using standard Taq polymerase results in little or no amplification in the enrichment PCR, for an organism with a critical primer mismatch there is little or no substrate for the standard sequencing primer in the sequencing reaction. Right column, percentage of P. acnes observed using such amplification and sequencing conditions.
D) Only when both a proofreading polymerase and a standard sequencing primer are used, are organisms with critical primer mismatches amplified and sequenced successfully. Right column, percentage of P. acnes observed using such amplification and sequencing conditions.
Supplementary Figure 6 Evidence of primer editing and differential recovery of an organism with mismatches to the V4 806R primer between the EMP (Taq) and DI (KAPA) methods.
A) Percent abundance of OTU 302446 as measured by either the EMP (Taq) or DI (KAPA) method.
B) Logo plots and alignments of V4 515F and V4 806R primer sequences to the corresponding region in reads assigned to OTU 302446 in the EMPCMB7 sample. Position with mismatch to the V4 806R primer is highlighted in red.
C) Percent abundance of the k__Bacteria;p__Tenericutes;c__Mollicutes;o__Anaeroplasmatales;f__Anaeroplasmataceae;g__ taxon as measured by either the EMP (Taq) or DI (KAPA) method.
D) Logo plots and alignments of V4 515F and V4 806R primer sequences to the corresponding region in reads assigned to the k__Bacteria;p__Tenericutes;c__Mollicutes;o__Anaeroplasmatales;f__Anaeroplasmataceae;g__ taxon in the EMPCMB7 sample. Position with mismatch to the V4 806R primer is highlighted in red.
Supplementary Figure 7 Evidence of primer editing and differential recovery of multiple taxa between the EMP (Taq) and DI (KAPA) methods in human samples.
A) Percent abundance of the k__Bacteria;p__TM7;c__TM7-3;o__CW040;f__F16;g__ taxon as measured by either the EMP (Taq) or DI (KAPA) method.
B) Logo plots and alignments of V4 515F and V4 806R primer sequences to the corresponding region in reads assigned to the k__Bacteria;p__TM7;c__TM7-3;o__CW040;f__F16;g__ taxon in the 7013.02.CF sample. Position with mismatch to the V4 515F primer is highlighted in red.
C) Percent abundance of the k__Bacteria;p__TM7;c__TM7-3;o__;f__;g__ taxon as measured by either the EMP (Taq) or DI (KAPA) method.
D) Logo plots and alignments of V4 515F and V4 806R primer sequences to the corresponding region in reads assigned to the k__Bacteria;p__TM7;c__TM7-3;o__;f__;g__ taxon in the 7000.01.CF sample. Position with mismatch to the V4 515F primer is highlighted in red.
E) Percent abundance of the k__Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Propionibacteriaceae;g__Propionibacterium taxon as measured by either the EMP (Taq) or DI (KAPA) method.
F) Logo plots and alignments of V4 515F and V4 806R primer sequences to the corresponding region in reads assigned to the k__Bacteria;p__Actinobacteria;c__Actinobacteria;o__Actinomycetales;f__Propionibacteriaceae;g__Propionibacterium taxon in the 6998.01.CF sample. Positions with mismatch to the V4 515F and V4 806R primers are highlighted in red.
Supplementary Figure 8 Recall and precision for individual data sets.
Recall and precision results of 1000 iterations of simulated re-noising by method for each dataset and comparison. For each pair of figures, the figure on the left represents the fraction of original differentiated taxa recovered by the respective method (recall) and the figure on the right represents the fraction of original differentiated taxa out of all (false positive and true positive) differentiated taxa by the respective method for a specific treatment comparison and dataset (precision).
Supplementary Figure 9 Comparison of shotgun and amplicon sequencing data.
A) Measured abundances of the even mock community by shotgun sequencing using 4 different library prep kits (Illumina Nextera XT (XT), KAPA Hyper Prep PCR (KP), KAPA Hyper Prep PCR-free (KF), and TrusSeq DNA PCR-free (TSF)), compared to qPCR genomic copy number data; from Jones et al. Since the mock community was pooled at an abundance of 5% per organism based on 16S rRNA gene copy number, the actual percent abundance of genomes per organism in this mock community can diverge from 5%. Jones et al3 attempted to determine genomic copy number by using organism-specific qPCR assays.
B) Root mean square deviation (RMSD) values for the HM-276D even mock community as determined by amplicon sequencing (data from Figure 1) or by shotgun sequencing (data from Jones et al).3 RMSD values for the amplicon data was calculated based on 5% per organism 16S rRNA gene abundance in the mock community. RMSD values for the shotgun data were calculated using the relative genomic abundance qPCR data from Jones et al.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Table 1 and Supplementary Notes 1–5 (PDF 3047 kb)
Rights and permissions
About this article
Cite this article
Gohl, D., Vangay, P., Garbe, J. et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol 34, 942–949 (2016). https://doi.org/10.1038/nbt.3601
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3601
This article is cited by
-
Bacteremia caused by Nocardia farcinica: a case report and literature review
BMC Infectious Diseases (2024)
-
Effect of ginger supplementation on the fecal microbiome in subjects with prior colorectal adenoma
Scientific Reports (2024)
-
Multi-factorial examination of amplicon sequencing workflows from sample preparation to bioinformatic analysis
BMC Microbiology (2023)
-
Activity budget and gut microbiota stability and flexibility across reproductive states in wild capuchin monkeys in a seasonal tropical dry forest
Animal Microbiome (2023)
-
In vitro gut microbiome response to carbohydrate supplementation is acutely affected by a sudden change in diet
BMC Microbiology (2023)