Abstract
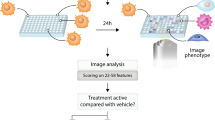
High-content, image-based screens enable the identification of compounds that induce cellular responses similar to those of known drugs but through different chemical structures or targets. A central challenge in designing phenotypic screens is choosing suitable imaging biomarkers. Here we present a method for systematically identifying optimal reporter cell lines for annotating compound libraries (ORACLs), whose phenotypic profiles most accurately classify a training set of known drugs. We generate a library of fluorescently tagged reporter cell lines, and let analytical criteria determine which among them—the ORACL—best classifies compounds into multiple, diverse drug classes. We demonstrate that an ORACL can functionally annotate large compound libraries across diverse drug classes in a single-pass screen and confirm high prediction accuracy by means of orthogonal, secondary validation assays. Our approach will increase the efficiency, scale and accuracy of phenotypic screens by maximizing their discriminatory power.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
van 't Veer, L.J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002).
Thomas, R.K. et al. High-throughput oncogene mutation profiling in human cancer. Nat. Genet. 39, 347–351 (2007).
Kolch, W. & Pitt, A. Functional proteomics to dissect tyrosine kinase signalling pathways in cancer. Nat. Rev. Cancer 10, 618–629 (2010).
Griffin, J.L. & Shockcor, J.P. Metabolic profiles of cancer cells. Nat. Rev. Cancer 4, 551–561 (2004).
Zhang, J., Yang, P.L. & Gray, N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009).
Kelloff, G.J. & Sigman, C.C. Cancer biomarkers: selecting the right drug for the right patient. Nat. Rev. Drug Discov. 11, 201–214 (2012).
Sundberg, S.A. High-throughput and ultra-high-throughput screening: solution- and cell-based approaches. Curr. Opin. Biotechnol. 11, 47–53 (2000).
Mayr, L.M. & Bojanic, D. Novel trends in high-throughput screening. Curr. Opin. Pharmacol. 9, 580–588 (2009).
Koehn, F.E. High impact technologies for natural products screening. Prog. Drug Res. 65 175, 177–210 (2008).
Lachance, H., Wetzel, S., Kumar, K. & Waldmann, H. Charting, navigating, and populating natural product chemical space for drug discovery. J. Med. Chem. 55, 5989–6001 (2012).
Nielsen, T.E. & Schreiber, S.L. Towards the optimal screening collection: a synthesis strategy. Angew. Chem. Int. Ed. Engl. 47, 48–56 (2008).
O' Connor, C.J., Beckmann, H.S. & Spring, D.R. Diversity-oriented synthesis: producing chemical tools for dissecting biology. Chem. Soc. Rev. 41, 4444–4456 (2012).
Caie, P.D. et al. High-content phenotypic profiling of drug response signatures across distinct cancer cells. Mol. Cancer Ther. 9, 1913–1926 (2010).
Klebe, G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discov. Today 11, 580–594 (2006).
Schneider, G. Virtual screening: an endless staircase? Nat. Rev. Drug Discov. 9, 273–276 (2010).
Inglese, J. et al. High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol. 3, 466–479 (2007).
Chen, B. et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat. Chem. Biol. 5, 100–107 (2009).
Wilson, C.J. et al. Identification of a small molecule that induces mitotic arrest using a simplified high-content screening assay and data analysis method. J. Biomol. Screen. 11, 21–28 (2006).
Perlman, Z.E. et al. Multidimensional drug profiling by automated microscopy. Science 306, 1194–1198 (2004).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Potts, M.B. et al. Using functional signature ontology (FUSION) to identify mechanisms of action for natural products. Sci. Signal. 6, ra90 (2013).
Young, D.W. et al. Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nat. Chem. Biol. 4, 59–68 (2008).
MacDonald, M.L. et al. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat. Chem. Biol. 2, 329–337 (2006).
Houle, D., Govindaraju, D.R. & Omholt, S. Phenomics: the next challenge. Nat. Rev. Genet. 11, 855–866 (2010).
Futamura, Y. et al. Morphobase, an encyclopedic cell morphology database, and its use for drug target identification. Chem. Biol. 19, 1620–1630 (2012).
King, K.R. et al. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip 7, 77–85 (2007).
Stegmaier, K. et al. Gene expression-based high-throughput screening(GE-HTS) and application to leukemia differentiation. Nat. Genet. 36, 257–263 (2004).
Kawatani, M. et al. Identification of a small-molecule inhibitor of DNA topoisomerase II by proteomic profiling. Chem. Biol. 18, 743–751 (2011).
Muroi, M. et al. Application of proteomic profiling based on 2D-DIGE for classification of compounds according to the mechanism of action. Chem. Biol. 17, 460–470 (2010).
Bantscheff, M. et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 (2007).
Feng, Y., Mitchison, T.J., Bender, A., Young, D.W. & Tallarico, J.A. Multi-parameter phenotypic profiling: using cellular effects to characterize small-molecule compounds. Nat. Rev. Drug Discov. 8, 567–578 (2009).
Roti, G. & Stegmaier, K. Genetic and proteomic approaches to identify cancer drug targets. Br. J. Cancer 106, 254–261 (2012).
Fuchs, F. et al. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol. Syst. Biol. 6, 370 (2010).
Bakal, C., Aach, J., Church, G. & Perrimon, N. Quantitative morphological signatures define local signaling networks regulating cell morphology. Science 316, 1753–1756 (2007).
Taylor, D.L. Past, present, and future of high content screening and the field of cellomics. Methods Mol. Biol. 356, 3–18 (2007).
Gustafsdottir, S.M. et al. Multiplex cytological profiling assay to measure diverse cellular states. PLoS One 8, e80999 (2013).
Wawer, M.J. et al. Toward performance-diverse small-molecule libraries for cell-based phenotypic screening using multiplexed high-dimensional profiling. Proc. Natl. Acad. Sci. USA 111, 10911–10916 (2014).
Cohen, A.A. et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511–1516 (2008).
Loo, L.H., Wu, L.F. & Altschuler, S.J. Image-based multivariate profiling of drug responses from single cells. Nat. Methods 4, 445–453 (2007).
Johnson, R.A. & Wichern, D.W. Applied Multivariate Statistical Analysis. 3rd edn. (Prentice Hall, Englewood Cliffs, N.J., 1992).
Martin, C.J. et al. Molecular characterization of macbecin as an Hsp90 inhibitor. J. Med. Chem. 51, 2853–2857 (2008).
Reddy, P. et al. Histone deacetylase inhibitor suberoylanilide hydroxamic acid reduces acute graft-versus-host disease and preserves graft-versus-leukemia effect. Proc. Natl. Acad. Sci. USA 101, 3921–3926 (2004).
Wójcik, C. et al. Valosin-containing protein (p97) is a regulator of endoplasmic reticulum stress and of the degradation of N-end rule and ubiquitin-fusion degradation pathway substrates in mammalian cells. Mol. Biol. Cell 17, 4606–4618 (2006).
Kuhn, D.J. et al. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood 110, 3281–3290 (2007).
Chen, D., Frezza, M., Schmitt, S., Kanwar, J. & Dou, Q.P. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr. Cancer Drug Targets 11, 239–253 (2011).
Kim, T.S. et al. Interaction of Hsp90 with ribosomal proteins protects from ubiquitination and proteasome-dependent degradation. Mol. Biol. Cell 17, 824–833 (2006).
Moffat, J.G., Rudolph, J. & Bailey, D. Phenotypic screening in cancer drug discovery- past, present and future. Nat. Rev. Drug Discov. 13, 588–602 (2014).
Kangas, J.D., Naik, A.W. & Murphy, R.F. Efficient discovery of responses of proteins to compounds using active learning. BMC Bioinformatics 15, 143 (2014).
Sigal, A. et al. Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nat. Methods 3, 525–531 (2006).
Schneider, C.A., Rasband, W.S. & Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Sugiyama, M. Dimensionality reduction of multimodal labeled data by local fisher discriminant analysis. J. Mach. Learn. Res. 8, 1027–1061 (2007)<>.
Schäfer, J. & Strimmer, K. A shrinkage approach to large-scale covariance matrix estimation and implications for functional genomics. Stat. Appl. Genet. Mol. Biol. 4, e32 (2005).
Wu, J., Hu, C.P., Gu, Q.H., Li, Y.P. & Song, M. Trichostatin A sensitizes cisplatin-resistant A549 cells to apoptosis by up-regulating death-associated protein kinase. Acta Pharmacol. Sin. 31, 93–101 (2010).
Ono, M. et al. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol. Cancer Ther. 3, 465–472 (2004).
Chen, M.C. et al. The HDAC inhibitor, MPT0E028, enhances erlotinib-induced cell death in EGFR-TKI-resistant NSCLC cells. Cell Death Dis. 4, e810 (2013).
Acknowledgements
We thank members of the Altschuler and Wu laboratories for critical feedback; U. Alon (Weizmann Institute of Science, Rehovot, Israel), and members of his laboratory for providing the CD tag plasmid and guidance on its use; G. DeMartino (University of Texas Southwestern Medical Center, Dallas) for useful conversations reagents for the proteasome validation and the Ub-R clone of HeLa cells; and S. Wei for help with HTS experiments. This research was partially supported by the US National Institutes of Health grants CA133253 (S.J.A.), R01CA184984 (L.F.W.) and the Institute of Computational Health Sciences at UCSF (S.J.A., L.F.W.).
Author information
Authors and Affiliations
Contributions
J.K., Q.W. and S.L. generated the reporter library; A.D.C. built pSEG; J.K. designed the experiments; J.K. and C.-H.H. performed the experiments; B.A.P. helped perform the HTS experiments; C.-H.H. performed the data analysis; J.K., C.-H.H., L.F.W. and S.J.A. wrote the manuscript; and L.F.W. and S.J.A. guided all aspects of this study.
Corresponding authors
Ethics declarations
Competing interests
S.J.A. and L.F.W. have submitted a patent application.
Integrated supplementary information
Supplementary Figure 1 Our reporter cell lines display diverse response to drugs from different functional classes.
Six reporter cell lines and the parent cell lines (untagged) were treated with various cancer drugs and imaged every 12 hrs for 48 hrs. Each row is a reporter cell line with the name of the YFP-tagged protein, and columns are drugs from 6 drug classes. Images at 48 hrs are shown. Blue: CFP. Green: YFP. Red: mCherry. The scale bar is 10 µm.
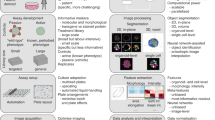
Supplementary Figure 2 Overview of profile computation.
1. Phenotypic features were extracted for each cell under DMSO control or drug/compound perturbations (e.g. Gemcitabine). 2. Drug/compound effects on each feature at population level (compared to DMSO) were summarized by KS statistics. 3. A phenotypic profile of a drug/compound (at single time point) consists of KS statistics of all features, and can be visualized as a point in high-dimensional feature space. 4. Phenotypic profiles can be extended to multiple time point. Dynamics of drug/compound effects were visualized as “time-traces” in the feature space.
Supplementary Figure 3 Phenotypic profiles show similarity within drug-class.
Phenotypic responses based on combining six different reporter cell lines (Supplementary figure 1) were computed. Each heat map summarizes phenotypic responses to one drug, and each row of 5 heat maps represents five different drugs in one drug class (e.g. DNA). To determine the y-axis sort order for each heat map, we: concatenated profiles from six reporters, sorted the features for camptothecin (CPT) from most green to most red, and then applied this sort order to all other drugs. The x-axis on each heat map is increasing time (0hr, 12hr,…, 48hr). Our figure shows that profiles are similar within drug classes (within each row of heat maps), and distinct between classes (across rows of heat maps), particularly at later times.
Supplementary Figure 4 Time traces of reporter-based phenotypic profiles.
The phenotypic profiles for six reporter cell lines (Supplementary figures 1, 3) were re-visualized as time-varying response curves in 3-D (and in stereo) using multidimensional scaling. Shown are phenotypic profiles for 30 drugs x 2 replicate experiments (solid lines and dash lines).
Supplementary Figure 5 Time points of 24 and/or 48 hrs are sufficient to discriminate drug classes.
Prediction accuracies were calculated using 10-fold cross-validation with phenotypic profiles built from the six reporter cell lines at 0hr only, 24hr only, 48hr only or all time points.
Supplementary Figure 6 Combining 24 and 48 hrs provides higher prediction accuracy.
Prediction accuracy was calculated using 10-fold cross validation for each of the 7 reference plates in the primary screen. The last column is the averaged prediction accuracy across the 7 reference plates.
Supplementary Figure 7 Prediction accuracy of NCI oncology set.
We calibrated confidence values for drug-class prediction using the NCI oncology set, which has “ground truth” annotation. As the confidence threshold increases (from blue to red; see color bar), the number of classified compounds decreases (x-axis). Accuracy (y-axis) was calculated as the proportion of classified compounds that were predicted correctly. The arrow indicates the confidence threshold = 0.1, which was used in our analysis.
Supplementary Figure 8 Statistics of primary hits across screened libraries.
A. Number of primary hits and hit rate in each library. B. Predicted drug classes for primary hits in each screened library.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 (PDF 1235 kb)
Supplementary Tables 1–6
Table 1 Feature list Table 2 Reference drug list Table 3 NCI oncology drug annotation and prediction Table 4 Literature supported compounds used as new reference drugs Table 5 Recall and new predictions Table 6 Validated hits and structure (XLSX 297 kb)
Rights and permissions
About this article
Cite this article
Kang, J., Hsu, CH., Wu, Q. et al. Improving drug discovery with high-content phenotypic screens by systematic selection of reporter cell lines. Nat Biotechnol 34, 70–77 (2016). https://doi.org/10.1038/nbt.3419
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3419
This article is cited by
-
High-content analysis identified synergistic drug interactions between INK128, an mTOR inhibitor, and HDAC inhibitors in a non-small cell lung cancer cell line
BMC Cancer (2024)
-
Genome-wide CRISPR screens identify ILF3 as a mediator of mTORC1-dependent amino acid sensing
Nature Cell Biology (2023)
-
Visual barcodes for clonal-multiplexing of live microscopy-based assays
Nature Communications (2022)
-
Image-based profiling for drug discovery: due for a machine-learning upgrade?
Nature Reviews Drug Discovery (2021)
-
Pharmaceutical therapeutics for articular regeneration and restoration: state-of-the-art technology for screening small molecular drugs
Cellular and Molecular Life Sciences (2021)