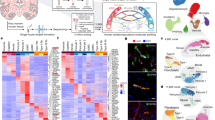
Abstract
Cerebrovascular disease is the third most common cause of death in developed countries, but our understanding of the cells that compose the cerebral vasculature is limited. Here, using vascular single-cell transcriptomics, we provide molecular definitions for the principal types of blood vascular and vessel-associated cells in the adult mouse brain. We uncover the transcriptional basis of the gradual phenotypic change (zonation) along the arteriovenous axis and reveal unexpected cell type differences: a seamless continuum for endothelial cells versus a punctuated continuum for mural cells. We also provide insight into pericyte organotypicity and define a population of perivascular fibroblast-like cells that are present on all vessel types except capillaries. Our work illustrates the power of single-cell transcriptomics to decode the higher organizational principles of a tissue and may provide the initial chapter in a molecular encyclopaedia of the mammalian vasculature.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
20 June 2018
In Fig. 1b of this Article, 'Csf1r' was misspelt 'Csfr1', and in Extended Data Fig. 11b, the genes in the first column had shifted down three rows. These errors have been corrected online.
References
Iadecola, C. The pathobiology of vascular dementia. Neuron 80, 844–866 (2013)
Zhao, Z., Nelson, A. R., Betsholtz, C. & Zlokovic, B. V. Establishment and dysfunction of the blood-brain barrier. Cell 163, 1064–1078 (2015)
Abbott, N. J., Rönnbäck, L. & Hansson, E. Astrocyte–endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 7, 41–53 (2006)
Zlokovic, B. V. The blood-brain barrier in health and chronic neurodegenerative disorders. Neuron 57, 178–201 (2008)
Phoenix, T. N. et al. Medulloblastoma genotype dictates blood brain barrier phenotype. Cancer Cell 29, 508–522 (2016)
de Lange, E. C. & Hammarlund-Udenaes, M. Translational aspects of blood-brain barrier transport and central nervous system effects of drugs: from discovery to patients. Clin. Pharmacol. Ther. 97, 380–394 (2015)
Majno, G. & Palade, G. E. Studies on inflammation. 1. The effect of histamine and serotonin on vascular permeability: an electron microscopic study. J. Biophys. Biochem. Cytol. 11, 571–605 (1961)
Simionescu, M., Simionescu, N. & Palade, G. E. Segmental differentiations of cell junctions in the vascular endothelium. The microvasculature. J. Cell Biol. 67, 863–885 (1975)
Ekman, N. et al. Bmx tyrosine kinase is specifically expressed in the endocardium and the endothelium of large arteries. Circulation 96, 1729–1732 (1997)
Wang, H. U., Chen, Z. F. & Anderson, D. J. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell 93, 741–753 (1998)
Hogan, B. M. et al. Vegfc/Flt4 signalling is suppressed by Dll4 in developing zebrafish intersegmental arteries. Development 136, 4001–4009 (2009)
Kutschera, S. et al. Differential endothelial transcriptomics identifies semaphorin 3G as a vascular class 3 semaphorin. Arterioscler. Thromb. Vasc. Biol. 31, 151–159 (2011)
Hirashima, M. & Suda, T. Differentiation of arterial and venous endothelial cells and vascular morphogenesis. Endothelium 13, 137–145 (2006)
Armulik, A., Genové, G. & Betsholtz, C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 21, 193–215 (2011)
Attwell, D., Mishra, A., Hall, C. N., O’Farrell, F. M. & Dalkara, T. What is a pericyte? J. Cereb. Blood Flow Metab. 36, 451–455 (2016)
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014)
Zeisel, A. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015)
Marques, S. et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352, 1326–1329 (2016)
Goldmann, T. et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17, 797–805 (2016)
Halpern, K. B. et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017)
Nguyen, L. N. et al. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature 509, 503–506 (2014)
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007)
van den Brink, S. C. et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017)
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014)
Iliff, J. J. et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid β. Sci. Transl. Med. 4, 147ra111 (2012)
Lindahl, P., Johansson, B. R., Levéen, P. & Betsholtz, C. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science 277, 242–245 (1997)
Armulik, A. et al. Pericytes regulate the blood-brain barrier. Nature 468, 557–561 (2010)
Daneman, R., Zhou, L., Kebede, A. A. & Barres, B. A. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature 468, 562–566 (2010)
Proebstl, D. et al. Pericytes support neutrophil subendothelial cell crawling and breaching of venular walls in vivo. J. Exp. Med. 209, 1219–1234 (2012)
Hall, C. N. et al. Capillary pericytes regulate cerebral blood flow in health and disease. Nature 508, 55–60 (2014)
Crisan, M. et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3, 301–313 (2008)
Göritz, C. et al. A pericyte origin of spinal cord scar tissue. Science 333, 238–242 (2011)
Soderblom, C. et al. Perivascular fibroblasts form the fibrotic scar after contusive spinal cord injury. J. Neurosci. 33, 13882–13887 (2013)
Hill, R. A. et al. Regional blood flow in the normal and ischemic brain is controlled by arteriolar smooth muscle cell contractility and not by capillary pericytes. Neuron 87, 95–110 (2015)
Guimaraes-Camboa, N. et al. Pericytes of multiple organs do not behave as mesenchymal stem cells in vivo. Cell Stem Cell 20, 345–359 (2017)
Nolan, D. J. et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev. Cell 26, 204–219 (2013)
Xie, L. et al. Sleep drives metabolite clearance from the adult brain. Science 342, 373–377 (2013)
Rajantie, I. et al. Bmx tyrosine kinase has a redundant function downstream of angiopoietin and vascular endothelial growth factor receptors in arterial endothelium. Mol. Cell. Biol. 21, 4647–4655 (2001)
He, L. et al. Analysis of the brain mural cell transcriptome. Sci. Rep. 6, 35108 (2016)
Vanlandewijck, M. et al. Primary isolation of vascular cells from murine brain for single cell sequencing. Protoc. Exch. https://doi.org/10.1038/protex.2017.159 (2018)
Vanlandewijck, M., Andrae, J., Gouveia, L. & Betsholtz, C. Isolation of vessel-associated pdgfra-H2BGFP positive cells from murine brain. Protoc. Exch. https://doi.org/10.1038/protex.2018.005 (2018)
Vanlandewijck, M., Andrae, J., Gouveia, L. & Betsholtz, C. Preparation of single cell suspensions from the adult mouse lung. Protoc. Exch. https://doi.org/10.1038/protex.2018.006 (2018)
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013)
Kwan, K. M. et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 (2007)
Asakawa, K. et al. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl Acad. Sci. USA 105, 1255–1260 (2008)
Fukuhara, S. et al. Visualizing the cell-cycle progression of endothelial cells in zebrafish. Dev. Biol. 393, 10–23 (2014)
Ando, K. et al. Clarification of mural cell coverage of vascular endothelial cells by live imaging of zebrafish. Development 143, 1328–1339 (2016)
Kikuchi, K. et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397–404 (2011)
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Ramsköld, D., Kavak, E. & Sandberg, R. How to analyze gene expression using RNA-sequencing data. Methods Mol. Biol. 802, 259–274 (2012)
Storvall, H., Ramskold, D. & Sandberg, R. Efficient and comprehensive representation of uniqueness for next-generation sequencing by minimum unique length analyses. PLoS One 8, e53822 (2013)
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010)
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995)
Grün, D. et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255 (2015)
Forrest, A. R. et al. A promoter-level mammalian expression atlas. Nature 507, 462–470 (2014)
Mackic, J. B. et al. Human blood-brain barrier receptors for Alzheimer’s amyloid-beta 1- 40. Asymmetrical binding, endocytosis, and transcytosis at the apical side of brain microvascular endothelial cell monolayer. J. Clin. Invest. 102, 734–743 (1998)
Do, T. M. et al. Oatp1a4 and an l-thyroxine-sensitive transporter mediate the mouse blood-brain barrier transport of amyloid-β peptide. J. Alzheimers Dis. 36, 555–561 (2013)
Deane, R. et al. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron 43, 333–344 (2004)
Bell, R. D. et al. SRF and myocardin regulate LRP-mediated amyloid-beta clearance in brain vascular cells. Nat. Cell Biol. 11, 143–153 (2009)
Xiong, H. et al. ABCG2 is upregulated in Alzheimer’s brain with cerebral amyloid angiopathy and may act as a gatekeeper at the blood-brain barrier for Aβ1–40 peptides. J. Neurosci. 29, 5463–5475 (2009)
Agarwal, S. et al. Active efflux of Dasatinib from the brain limits efficacy against murine glioblastoma: broad implications for the clinical use of molecularly targeted agents. Mol. Cancer Ther. 11, 2183–2192 (2012)
Lam, F. C. et al. β-Amyloid efflux mediated by p-glycoprotein. J. Neurochem. 76, 1121–1128 (2001)
Corder, E. H. et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science 261, 921–923 (1993)
Tarlungeanu, D. C. et al. Impaired amino acid transport at the blood brain barrier is a cause of autism spectrum disorder. Cell 167, 1481–1494 (2016)
Dieterich, L. C. et al. Transcriptional profiling of human glioblastoma vessels indicates a key role of VEGF-A and TGFβ2 in vascular abnormalization. J. Pathol. 228, 378–390 (2012)
Acknowledgements
This study was supported by AstraZeneca AB (C.B., U.L.), the Swedish Research Council (C.B.: 2015-00550; U.L.: K2014-64X-20097-09-5), the European Research Council (C.B.: AdG294556), the Leducq Foundation (C.B., A.K.: 14CVD02), Swedish Cancer Society (C.B.:150735; U.L.:CAN 2016/271), Knut and Alice Wallenberg Foundation (C.B.: 2015.0030), Hjärnfonden (U.L.), Swiss National Science Foundation (A.K.: 31003A_159514/1) and the Synapsis Foundation (A.K.). We thank C. Olsson, H. Leksell, P. Peterson, J. Chmielniakova, K. Gaengel, BioVis (Uppsala), Center for Microscopy and Image Analysis (Zurich) and Eukaryotic Single Cell Genomics facility (Science for Life Laboratory) for technical help, and K. Alitalo, P. Soriano, D. Silver and L. Sorokin for reagents.
Author information
Authors and Affiliations
Contributions
M.V., L.H., N.M., U.L. and C.B. conceived and designed the project; M.V., M.A.M., J.A., K.A., F.D.G., K.N., T.L., B.L., E.R., L.G., Y.Z., M.R., A.K. and C.B. performed experiments; L.H. performed bioinformatic analysis; L.H. and Y.S. constructed the online database; C.B., L.H. and M.V. analysed the bioinformatic data; C.B. and U.L. wrote the manuscript with substantial input from M.V., L.H., M.A.M., J.A. and K.A. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks D. McDonald and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Cell sorting, clustering and annotation and control for batch effects.
a, Representative FACS plots for cell sorts from the various transgenic reporter lines and C57Bl6 controls. For all sorts, the cells were first roughly selected with a broad gate based on forward scatter (FSC)/side scatter (SSC). This initial gate was then used as parent gate for cell selection based upon the relevant fluorescent signal of the different reporters. No additional gating was included, and the cut-off between positive and negative fluorescent signals was assessed using non-fluorescent C57Bl6 mice. b, BackSPIN analysis of cells obtained from Cldn5-GFP, Pdgfrb-GFP;Cspg4-DsRed and Pdgfra-H2BGFP mice. The BackSPIN algorithm splits the cells stepwise and also assigns genes uniquely to one branch with each split. At split level 1, all endothelial cells were assigned to the right branch, whereas all other cell types were assigned to the left branch. BackSPIN continues to split for as long as it recognizes expression heterogeneity among the genes assigned to that cluster leading to that some clusters continue to split dichotomously, whereas other clusters stop splitting at a certain level. Cluster 0, containing pericytes, stopped splitting at level 2. Any level can be arbitrarily chosen for cluster annotation; we chose level 6, at which 38 clusters had formed. c, The expression of the housekeeping gene Actb across the 38 clusters in b is illustrated as a bar plot, where each bar represents one cell (∼3,500 in total). A gross annotation of the 38 clusters based on cell class-specific marker expression (see Fig. 1 for examples) is depicted below the bar plot. d, Abbreviations for the cell types. e, A reanalysis of cluster 0 from b, c by BackSPIN using all genes generated dichotomous splits over six levels, showing that this cluster had stopped spitting in b because differentially expressed genes had been assigned to other branches. f, In the 64 clusters generated in e, expression of SMC markers, such as Acta2, was found in the right-most clusters. These clusters were subsequently identified as vSMC. g, The same tree as in b, with cluster 0 split in to PC and vSMC at level 7. Some clusters at level 6 appeared similar and were consolidated to a higher node, as indicated by the red boxes. h, The resulting simplified tree, which also shows the final cell type annotation that is used to display the data throughout the paper and at http://betsholtzlab.org/VascularSingleCells/database.html. i, An excerpt from the database illustrates the expression of Actb across the cell types. Cluster 1 at level 6 in b, g contained four cells displaying astrocyte markers; this cluster was considered too small and was removed from the dataset. Astrocytes were instead obtained from Tagln-Cre;R26-stop-tdTomato mice and added to the dataset for comparison. j, The position of the major cell classes in a t-SNE diagram, as annotated using marker expression. k, Representative marker distribution for the cell classes in t-SNE diagrams. l, m, Control for batch effects. Neither t-SNE nor SPIN showed any clustering bias related to mouse individual or experimental repetitions. l, The distribution of 1,385 mural cells colour-coded for their mouse origin in t-SNE (top) and SPIN diagrams (bottom: an arbitrarily chosen pan-mural cell marker Myl9 is shown). m, The distribution of 1,100 endothelial cells in t-SNE (top) and SPIN (bottom: the zonated endothelial-specific transcript Mfsd2a is shown).
Extended Data Figure 2 Average expression and cell sub-types.
a, Average level of expression in the different cell types of the markers shown in Fig. 1b. A key to the cell type abbreviations is provided in Extended Data Fig. 1. Diagrams of the same type for all genes are available at http://betsholtzlab.org/VascularSingleCells/database.html. b, Microglial subtypes. The microglial markers Cx3cr1 and Mrc1 both mark the microglial (MG) cluster, as shown in the bar-plot diagrams (left) and in the average diagram (middle). Right: insets show magnified views of bar-plots indicated by black arrows. Of the 14 microglial cells present in the MG cluster, the eight left-most cells express Cx3cr1 but not Mrc1, whereas the six right-most cells show the reverse pattern. Since Cx3cr1 is a marker for interstitial microglia and Mrc1 is a marker for perivascular microglia and macrophages, this pattern suggests that both these subtypes of microglia are present in the MG cluster, and that they are separated by SPIN to opposite ends of the cluster. c, Oligodendrocyte (OL) subtypes. The marker for differentiating oligodendrocytes Opalin is expressed in the 150 right-most out of the 155 cells in the OL cluster, whereas the oligodendrocyte precursor cell (OPC) marker Pdgfra is expressed only in the five left-most cells (note that Pdgfra is also highly expressed in the FB1 and FB2 clusters). Since the OLs were sorted from an initial preparation of brain vascular fragments (in turn isolated using anti-PECAM1 coated magnetic beads) from Pdgfra-H2BGFP mice, the Opalin-positive cells appear physically associated with blood vessels. As for the microglial subtypes, SPIN separates the two oligodendrocyte lineage cell subtypes to opposite ends of the cluster. d, Subtypes of fibroblast-like cells. Numerous markers distinguish cells in the FB1 and FB2 clusters. Here we show the examples Col15a1, which is expressed preferentially in FB1 cells, and Itga8, which is expressed preferentially in FB2 cells.
Extended Data Figure 3 Endothelial arteriovenous zonation of Bmx, MFSD2A, TFRC and SLC16A1.
All panels show microscopic fields from cerebral cortex with different labels for endothelial cells (cyan: PECAM1), mural cells including SMCs and pericytes (white: PDGFRB or DES), arterial SMCs (green: ACTA2), specific zonated proteins (red: MFSD2A, TFRC or SLC16A1), separately, or in the indicated combinations. A, artery or arteriole; V, vein or venule; Cap, capillary/ies. a, Validation of Bmx as an arterial marker in adult mouse brain using Bmx-lacZ mice. ACTA2 immunofluorescence distinguishes arteries from veins and capillaries. PECAM1 immunofluorescence depicts all vessels. b–d, The first row of each panel shows the same microscopic field at overview magnification for different immunofluorescence stainings. Hatched boxes delineate the areas shown at higher magnifications in the second row of each panel. The boxes are individually labelled i–vi. Arteries and arterioles are identified by ACTA2. Veins and venules are identified by vessel diameter in combination with the absence of ACTA2. Capillaries are identified by diameter and association with PDGFRB or DES-positive pericytes. An alternative choice of PDGFRB or DES labelling for mural cells was made to match the co-staining with other antibodies. b, Zonated expression of MFSD2A. Note the strong expression in capillaries and weak or absent staining in arteries/arterioles and veins. c, Zonated expression of TFRC. Note the strong expression in capillaries and veins/venules but absent staining in arteries/arterioles. d, Zonated expression of SLC16A1. Note the strong expression in capillaries and veins/venules but absent expression in arteries/arterioles. Data were reproduced in sections taken from three different mice. Scale bars, 20 μm.
Extended Data Figure 4 Endothelial arteriovenous zonation of VCAM1, VWF, Gkn3 and Slc38a5.
All panels show data from cerebral cortex. a, b, Immunofluorescence with different labels for endothelial cells (cyan, PECAM1 or PODXL), mural cells including SMC and pericytes (white, PDGFRB), arterial SMCs (green, ACTA2), and specific zonated proteins (red, VCAM1 or VWF), separately, or in the indicated combinations. The first row of each panel of a, b shows the same microscopic field at overview magnification for different immunofluorescent stainings. Hatched boxes delineate the areas shown at higher magnifications in the second row of each panel. The boxes are individually labelled i–vi. Arteries and arterioles (A) are identified by ACTA2. Veins and venules are identified by vessel diameter in combination with the absence of ACTA2. Capillaries are identified either by TFRC-positive staining and diameter, or by diameter and association with PDGFRB-positive pericytes. a, Zonated expression of VCAM1. Note the strong expression in veins, weak but clear expression in arteries/arterioles, and absent expression in capillaries. b, Zonated expression of VWF. Note the strong expression in arteries and veins, weak expression in arterioles and venules and weak expression in capillaries. c, d, RNA ISH for the indicated genes validates the zonal distribution of Gkn3 to arteries and Slc38a5 to veins. e, Quantification of the ISH results. The following number of mice and fields in total were analysed: Slc38a5 (n = 3, 15 fields); Gkn3 (n = 2, 20 fields). In Fiji software the ROI manager was used to mark and measure the mean grey values of the vascular segments of interest (vein, artery) and data were presented using GraphPad Prism 6 software. An unpaired t-test with Welsh correction was used to demonstrate statistical significance in gene expression between arteries and veins. x-axis shows the vascular segment, y-axis shows the mean grey values. Unpaired t-test with Welsh correction demonstrates statistically significant difference for both genes (****P < 0.0001, ***P < 0.001). Data were reproduced in sections taken from three different mice. Scale bars, 20 μm.
Extended Data Figure 5 Validation of endothelial arteriovenous zonated gene expression using Allen Brain Atlas.
Allen Brain Atlas data for nine zonated endothelial transcripts (Gkn3, Vwf, Slc38a5, Slc6a6, Slc7a5, Mfsd2a, Slc16a1, Vcam1 and Mgp) and one highly expressed endothelial transcript that does not show statistically significant zonation (Cldn5). The left panel of diagrams shows the expression distribution across all vascular cell types clustered by BackSPIN and depicted by average expression for each cluster. These diagrams are excerpts from http://betsholtzlab.org/VascularSingleCells/database.html, where bar plots can also be accessed and viewed at high magnification. A key to the cell type abbreviations is provided in Extended Data Fig. 1. The middle panel of diagrams shows bar-plots for 1,100 endothelial cells (the clusters indicated by the black arrow) organized in one dimension by SPIN. The grey bars show the expression level in individual cells and the red curve (loess function) depicts the expression pattern across the range. The arteriovenous zonation indicated above the middle panel of diagrams is suggested by the data shown in Fig. 2 and Extended Data Figs 3, 4: venous endothelial cells to the left, capillary in the middle and arterial cells to the right. The right panel of micrographs shows excerpts from the Allen Brain Atlas, one overview and two at higher magnification (black frames). Genes with preferential expression in arteries (for example, Gkn3, Mgp), veins (for example, Slc38a5) or both (for example, Vwf, Vcam1 and Slc6a6) show distinct signals in large penetrating as well as in deep vascular structures. Genes that are highly expressed in capillaries (Mfsd2a, Slc16a1, and Cldn5) show distinct labelling of small vessel structures at all locations. The strong expression of Vcam1 and Mgp in fibroblast-like cells may contribute to the labelling of large vessels, although immunofluorescence shows a preferential localization of Vcam1 to endothelial cells (Fig. 3). Red arrows point to labelled profiles consistent with arteries/arterioles or veins. Red arrowheads point to profiles consistent with microvessels. Allen Brain Atlas experiment codes are provided above each overview image. Scale bars, 200 μm.
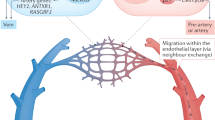
Extended Data Figure 6 Zonation principles, endothelial transcription factors and transporters, and genes specific to mural cell subtypes.
a, Schematic illustration of zonation principles and distribution of cell types. Endothelial arteriovenous zonation: red–blue; mural arteriovenous zonation: green–yellow; fibroblast-like cells and LAMA1 sleeve: brown. Arrow, arteriolar-capillary boundary. b, c, Heat map of relative expression (red, high; blue, low) of 120 transcription factor (b) and 145 transporter (c) transcripts across the SPIN range of 1,100 endothelial cells. Venous (V), capillary (Cap) and arterial (A) position in the range is depicted at the top. The identities of the transcripts distributing according to six or five major patterns of zonation, respectively, are provided to the right of the heat maps. d, Morphometric analysis of vessel diameter at the transitions between cells positive (+) or negative (−) for immunofluorescent labelling of the indicated proteins. The following number of mice and vessel profiles were analysed for each value: diameter at LAMA1 +/− transition, 5 mice, 49 vessels; diameter at TAGLN/ACTA2 +/− transition, 3 mice, 66 vessels; diameter at CNN1+/− transition, 3 mice, 34 vessels. e, Heat map of relative expression (red, high; blue, low) of the transcripts most specific for each subtype of mural cells ordered according to anatomical axis (vSMC, PC, aaSMC and aSMC). Transcript identities for each group are provided to the right of the heat map. f, Immunofluorescence shows abrupt transition from arteriolar SMCs to pericytes (arrows). Scale bars, 50 μm.
Extended Data Figure 7 Zonation of mural cell markers in mouse brain.
a, b, Immunofluorescence with different labels for endothelial cells (cyan, PECAM1), and mural cells including SMCs and pericytes (white, PDGFRB), arterial SMC (green, ACTA2), specific zonated proteins/reporters (red, TAGLN or CSPG4–DsRed) or in the indicated combinations. All images are from cerebral cortex. These panels show protein distribution to vessel types that match the zonal distribution of transcripts to mural cell subtypes, as shown in Fig. 3a. Data were reproduced in sections taken from three different mice. Scale bars, 50 μm. c, d, Heat map of relative expression (red, high; blue, low) of 131 transcription factor (c) and 121 transporter (d) transcripts across the SPIN range of 1,385 mural cells. The anatomical positions (capillary (Cap), venous (V), arteriolar (Aa) and arterial (A)) are shown at the top. Note the presence of two zones and their non-alignment with the anatomical axis (punctuated zonation where arterioles and veins meet). The identity of the transcripts distributing according to the two major patterns of zonation (SMC-high and PC-high) is provided to the right. e, Examples of three arterial SMC transcript distribution patterns exemplified by Cnn1, Tinagl1 and Fos and other transcripts with similar distribution. The transcripts with Fos-like distribution include products of numerous immediate early genes (IEGs), including Egr1, Fosb, Jun and Junb.
Extended Data Figure 8 Zonation of mural cell markers in zebrafish brain.
a, The zonated distribution of tagln and pdgfrb as shown in the three-month-old zebrafish brain using transgenic reporter fish. GFP is expressed from the tagln promoter and RFP from the UAS promoter activated by pdgfrb promoter-driven Gal4FF. Similar to mice, tagln is expressed in a/aaSMCs (white arrows, inset i) whereas pdgfrb marks mural cells along all vessel types, including capillary pericytes (PC). The lower panels show insets at high magnification. Note the lower expression of pdgfrb in the aSMCs of the larger arteries (inset ii). b, GFP expression from a UAS promoter activated by Gal4FF expressed from the abcc9 promoter in forebrain mural cells of a two-month-old zebrafish. Endothelial cells are labelled by DsRed expression from the kdrl (Vegfr2) promoter. Note that, similar to the expression pattern of Abcc9 in mice, fish abcc9 expression is localized mainly to capillary pericytes and venous mural cells, but not to arteries or arterioles (A). Top, a midsagittal view shows GFP-positive pericytes (white arrowheads) on capillary branches, whereas the SMC of the feeding arteriole as well as the upstream artery (A, white arrows) are GFP-negative. Middle, in a ventral view of the capillary network branching off an arteriole (A), numerous GFP-positive pericytes are seen, while the arteriole is negative. An arteriole–capillary transition is indicated by a yellow arrowhead. Dashed line indicates a vascular continuum stretching from the arteriole (A) across capillary branches to a venule (V). Bottom, another midsagittal view illustrates that arterial SMCs (A) lack abcc9 expression. Scale bars, 50 μm.
Extended Data Figure 9 Pericyte definition, organotypicity and transporter transcript expression.
a, Bar plot showing the expression of the highly abundant endothelial marker Cldn5 across the BackSPIN clusters; note the abundant and specific expression in EC clusters. The arrow points to a small group of cells in the PC cluster showing Cldn5 expression. The BackSPIN range of 1,088 pericytes is shown at larger magnification in the lower panels. An additional seven high abundant endothelial markers (Pecam1, Ly6a, Ly6c1, Podxl, Ptprb, Slc2a1 and Slco1c1) all show a peak of expression in the same roughly 10 cells, suggesting endothelial contamination of these pericytes. The high abundance markers for fibroblast-like cells (Lum) and astrocytes (Aqp4) do not reveal signs of contamination of the pericytes by these cell types. b, Annotation of lung vascular cell BackSPIN clusters and identification of lung pericytes. The PC cluster was identified according to the expression of known and/or shared markers with brain pericytes, combined with the absence of markers for vascular smooth muscle cells (VSMC), fibroblasts (FB), chondrocytes/perichondrium (CP), or endothelial cells (EC). The mixed cluster contains markers for epithelial cells together with markers for fibroblasts and probably represents contaminated cells. Among the endothelial cells (EC), several clusters were identified, including arterial (a), capillary (c/capil) and lymphatic (LEC). c, t-SNE analysis of brain and lung cells shows separation of brain and lung pericytes, as well as of brain and lung fibroblast-like cells, into distinct clusters suggesting organotypic differences for both cell classes. d, Bar plot of expression of the transporter transcript Atp13a5 in brain shows that it is highly specific for pericytes and vSMCs. e, Heat map illustrating the expression (red, high; blue, low) of the 200 most variable transporter transcripts across the brain vascular and vessel-associated cell types. Hierarchical clustering divides the patterns of transporter distribution into approximately 13 groups (separated by hatched lines). The identity of the transcripts in each group is provided to the right of the heat map. Note the abundance of transporters that are pericyte-specific.
Extended Data Figure 10 Localization of perivascular fibroblast-like cells.
a, Pdgfra-H2BGFP-positive cells in veins. Right panels show that the H2BGFP-positive nuclei are distinct from Cspg4-DsRed- and Pdgfrb-GFP-positive mural cells and GFAP-positive astrocytes. White arrows indicate Pdgfra-H2BGFP-positive nuclei. b, Pdgfra-H2BGFP-positive nuclei associated with venules are located between the endothelial cells and AQP4-positive astrocyte end-feet. Capillary-associated Pdgfra-H2BGFP-positive nuclei are located outside the astrocyte end-feet (insets); these cells probably represent oligodendrocyte lineage cells. c, The distribution of LAMA1 correlates with the distribution of vessel-associated Pdgfra-H2BGFP-positive cells. Image in c is from striatum and images in a and b are from cerebral cortex. Data were reproduced in sections taken from three different mice. Scale bars, 20 μm.
Extended Data Figure 11 Characterization of fibroblast-like cells in the brain.
a, GO analysis of the 50 most specific transcripts for the following four cell categories: fibroblast-like cells (FB1, FB2), pericytes and venous SMCs (PC, vSMC), arteriolar and arterial SMCs (aaSMC, aSMC) and endothelial cells (EC). The transcripts were selected on the basis of their distribution correlation with Pdgfra, Abcc9, Acta2 and Pecam1, respectively. Right panel shows a selection of significant terms for each cell class in the three mentioned categories. b, Further categorization of the 50 top markers for fibroblast-like cells reveals that most of them are associated with ECM components, turnover, or function. A few are related to innate immunity function. Nearly all markers are also expressed by lung fibroblasts. c, d, Expression heat map (blue, low; red, high) of collagen transcripts in mural cells and fibroblast-like cells. c, Absolute expression. d, Relative expression. Note the abundance of collagen transcripts in the fibroblast-like cell clusters (FB1 and FB2) and the difference between them, indicating FB subtypes. e, Bar plot distribution of Lum shows strong specificity for the FB clusters. Immunofluorescence for LUM shows its specific localization to blood vessels larger than capillaries and a correlation with the distribution of Pdgfra–H2BGFP-positive perivascular cells. f, Bar plot distribution of Dcn shows strong specificity for the FB clusters. Allen Brain Atlas images confirm expression in large penetrating brain vessels (red arrows).
Extended Data Figure 12 Vascular cell type-specific expression of genes linked to brain diseases.
Brain vascular scRNA-seq data provide insight into brain disease mechanisms. Three types of disease mechanisms are illustrated. For genes implicated in the transport of Alzheimer’s amyloid-β (Aβ) peptide across the BBB, it is noteworthy that several different cell types of the neurovascular unit are implicated in both Aβ influx and efflux mechanisms. Surprisingly, Lrp1 shows its highest expression in perivascular fibroblast-like cells (FB) and pericytes (PC) or vSMCs, but no detectable expression in endothelial cells (EC). *OMIM and #MIM numbers are provided for the monogenic brain disorders. We list the four genes known to be mutated in primary familial brain calcification (PFBC) to illustrate the different cellular expression of these genes, suggesting that changes in cell–cell communication in the vascular wall plays a role in the pathogenesis of this disease. It is noteworthy that several of the genes mutated in other listed monogenic brain disorders are commonly expressed in the cell types molecularly defined through the present study, such as Srpx2 (FB) and Uchl1 (PC). Other transcripts, including Slc2a1 and Mfsd2a, show the expected expression in EC. Unexpectedly, Jam3, which is thought to be expressed mainly in EC, has its major vascular expression in mural cells and oligodendrocytes (OL). Of genes overexpressed in glioblastoma grade IV (GBM) vessels, it is striking that most have their normal expression site in FB and PC or vSMC. This may suggest that GBM vasculature has an altered cellular composition, including a higher abundance of mural and perivascular cells, possibly reflecting a fibrotic reaction originating from FB. The diagrams display relative levels of expression. High-resolution quantitative versions of the diagrams and single cell bar plots can be viewed at http://betsholtzlab.org/VascularSingleCells/database.html. **A detailed analysis of the expression of the microglial expression of Apoe shows that it is predominantly expressed in perivascular brain macrophages.
Supplementary information
Supplementary Table 1
This file contains endothelial specific zonated genes.
Supplementary Table 2
This file contains the top 500 endothelial specific differentially expressed genes.
Supplementary Table 3
This file shows differential expression of mural cell specific genes.
Supplementary Table 4
This file contains a list of used antibodies.
Localization of perivascular fibroblast-like cells
A z-stack video taken perpendicular to the midsagittal plane visualizes the location of the pdgfra-H2BGFP positive perivascular cells. Note that, on larger vessels, the cells are inside of the AQP4-positive astrocyte end-feet (Red: AQP4), but outside of the vessel (White: CD31). Capillary-associated pdgfra-H2BGFP positive cells likely represent cells from the oligodendrocyte lineage, and are localized outside of the astrocyte end-feet. See Extended Data Figure 11b for high-resolution still images of the video.
Rights and permissions
About this article
Cite this article
Vanlandewijck, M., He, L., Mäe, M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). https://doi.org/10.1038/nature25739
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25739
This article is cited by
-
Analysis of ischemic stroke-mediated effects on blood–brain barrier properties along the arteriovenous axis assessed by intravital two-photon imaging
Fluids and Barriers of the CNS (2024)
-
Blood–brain borders: a proposal to address limitations of historical blood–brain barrier terminology
Fluids and Barriers of the CNS (2024)
-
Platelet-derived growth factor signaling in pericytes promotes hypothalamic inflammation and obesity
Molecular Medicine (2024)
-
Fasting upregulates the monocarboxylate transporter MCT1 at the rat blood-brain barrier through PPAR δ activation
Fluids and Barriers of the CNS (2024)
-
Endothelial and mural laminin-α5 contributes to neurovascular integrity maintenance
Fluids and Barriers of the CNS (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.