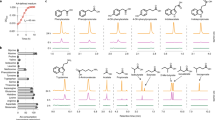
Abstract
The human gut microbiota produces dozens of metabolites that accumulate in the bloodstream1,2, where they can have systemic effects on the host. Although these small molecules commonly reach concentrations similar to those achieved by pharmaceutical agents, remarkably little is known about the microbial metabolic pathways that produce them. Here we use a combination of genetics and metabolic profiling to characterize a pathway from the gut symbiont Clostridium sporogenes that generates aromatic amino acid metabolites. Our results reveal that this pathway produces twelve compounds, nine of which are known to accumulate in host serum. All three aromatic amino acids (tryptophan, phenylalanine and tyrosine) serve as substrates for the pathway, and it involves branching and alternative reductases for specific intermediates. By genetically manipulating C. sporogenes, we modulate serum levels of these metabolites in gnotobiotic mice, and show that in turn this affects intestinal permeability and systemic immunity. This work has the potential to provide the basis of a systematic effort to engineer the molecular output of the gut bacterial community.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nicholson, J. K. et al. Host–gut microbiota metabolic interactions. Science 336, 1262–1267 (2012)
Wikoff, W. R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl Acad. Sci. USA 106, 3698–3703 (2009)
Danaceau, J. P., Anderson, G. M., McMahon, W. M. & Crouch, D. J. A liquid chromatographic–tandem mass spectrometric method for the analysis of serotonin and related indoles in human whole blood. J. Anal. Toxicol. 27, 440–444 (2003)
Venkatesh, M. et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41, 296–310 (2014)
Chyan, Y. J. et al. Potent neuroprotective properties against the Alzheimer β-amyloid by an endogenous melatonin-related indole structure, indole-3-propionic acid. J. Biol. Chem. 274, 21937–21942 (1999)
Elsden, S. R., Hilton, M. G. & Waller, J. M. The end products of the metabolism of aromatic amino acids by clostridia. Arch. Microbiol. 107, 283–288 (1976)
Dickert, S., Pierik, A. J., Linder, D. & Buckel, W. The involvement of coenzyme A esters in the dehydration of (R)-phenyllactate to (E)-cinnamate by Clostridium sporogenes. FEBS J. 267, 3874–3884 (2000)
Dickert, S., Pierik, A. J. & Buckel, W. Molecular characterization of phenyllactate dehydratase and its initiator from Clostridium sporogenes. Mol. Microbiol. 44, 49–60 (2002)
Heap, J. T. et al. The ClosTron: mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 80, 49–55 (2010)
Lovitt, R. W., Morris, J. G. & Kell, D. B. The growth and nutrition of Clostridium sporogenes NCIB 8053 in defined media. J. Appl. Bacteriol. 62, 71–80 (1987)
Su, L. et al. Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology 136, 551–563 (2009)
Bendall, S. C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011)
Theys, J. et al. Repeated cycles of Clostridium-directed enzyme prodrug therapy result in sustained antitumour effects in vivo. Br. J. Cancer 95, 1212–1219 (2006)
Bader, J. & Simon, H. ATP formation is coupled to the hydrogenation of 2-enoates in Clostridium sporogenes. FEMS Microbiol. Lett. 20, 171–175 (1983)
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014)
Nurk, S. et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737 (2013)
Aziz, R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75 (2008)
Medema, M. H., Takano, E. & Breitling, R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 30, 1218–1223 (2013)
Case, R. J. et al. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl. Environ. Microbiol. 73, 278–288 (2007)
Sonnenburg, E. D. et al. Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252 (2010)
Martens, E. C., Chiang, H. C. & Gordon, J. I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 (2008)
Spitzer, M. H. et al. An interactive reference framework for modeling a dynamic immune system. Science 349, 1259425 (2015)
Finck, R. et al. Normalization of mass cytometry data with bead standards. Cytometry A 83A, 483–494 (2013)
Elder, B. L., Boraker, D. K. & Fives-Taylor, P. M. Whole-bacterial cell enzyme-linked immunosorbent assay for Streptococcus sanguis fimbrial antigens. J. Clin. Microbiol. 16, 141–144 (1982)
Bader, J., Rauschenbach, P. & Simon, H. On a hitherto unknown fermentation path of several amino acids by proteolytic clostridia. FEBS Lett. 140, 67–72 (1982)
Bühler, M., Giesel, H., Tischer, W. & Simon, H. Occurrence and the possible physiological role of 2-enoate reductases. FEBS Lett. 109, 244–246 (1980)
Acknowledgements
We thank A. I. Scott for technical assistance and S. Yoshida (Kyoto University) for critical review of the manuscript. This work was funded by a grant from the National Institutes of Health NIDDK (R01-DK101674) to J.L.S. and M.A.F., an NIH Director’s New Innovator Award (DP2-OD006515) to J.L.S., and Early Independence Award (DP5-OD023056) to M.H.S., FDA Grant BAA-12-00118 to G.P.N., NIH Awards U19AI057229, U19AI100627, R33CA183654, R33CA0183692, R01GM10983601, R01CA184968, R01CA19665701, R21CA183660, R01NS08953301, 5UH2AR067676 and R01HL120724 to G.P.N., Department of Defence Grants OC110674 and W81XWH-14-1-0 180 to G.P.N., Gates Foundation Grant OPP1113682 to G.P.N., DP1 DK113598 to M.A.F., R01 DK110174 to M.A.F., HHMI-Simons Faculty Scholars Award to M.A.F., Byers Award in Basic Science to M.A.F., a Fellowship for Science and Engineering from the David and Lucile Packard Foundation to M.A.F. and a BASF research grant to M.A.F, and two Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Awards to M.A.F. and J.L.S. A.J.H. was supported by an NIH postdoctoral NRSA (T32-AI007328). W.V.T. and B.D.M. were each supported by a National Science Foundation Graduate Research Fellowship Grant No. DGE-114747.
Author information
Authors and Affiliations
Contributions
D.D., M.A.F. and J.L.S. conceived the study. D.D. generated the mutant bacteria. D.D., A.J.H. and W.V.T. performed the anaerobic growth curve experiments. A.L. and T.M.C. supervised generation of the LC–MS/MS assay. D.D. performed the metabolite incubations and LC–MS/MS experiments. B.D.M. assembled genomes from publicly available raw sequence data. D.D. and B.D.M. analysed genomic data. D.D. and S.K.H. performed gnotobiotic experiments. M.H.S. performed mass cytometry experiments and analysed data. G.P.N. supervised mass cytometry experiments and analysis. D.D., M.A.F. and J.L.S. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks M. Blaut, M. Redinbo, G. Siuzdak and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Search for an indolelactate dehydratase.
a, Phylogenetic analyses indicate that FldC (CLOSPO_00311) is distinct from its closest isospecific, heterofunctional homologue (HadC, CLOSPO_02758) in C. sporogenes. The neighbour-joining tree was built from the top 100 BLAST hits to FldC from the GenBank database. Sequences from very similar species and strains are collapsed into red branches. b, Genomic context for fldC and the most similar gene (hadC) in C. sporogenes. The amino acid sequence similarity between FldC and HadC is 47%.
Extended Data Figure 2 Generation of the phenyllactate dehydratase disruption mutant.
a, The ClosTron bacterial type II intron system was used to disrupt fldC (CLOSPO_00311) by insertion of an erythromycin-containing intron within the coding sequence at nucleotide position 561. b, Successful integration at the fldC locus was determined by amplifying DNA using primers flanking the region of insertion. The expected PCR product for the wild type is 600 bp as shown in the ethidium-bromide-stained agarose gel from a single experiment. Successful chromosomal integration was confirmed by Sanger sequencing.
Extended Data Figure 3 Summary of biochemical pathways for AAA metabolism by C. sporogenes.
Phenylalanine, tyrosine and tryptophan are all metabolized through the reductive pathway by the same enzymes. The first step is an aminotransferase reaction, probably catalysed by aromatic amino acid aminotransferase (Aat). This enzyme activity has been demonstrated in C. sporogenes cells, however, the gene encoding this enzyme has not been identified. The arylpyruvates are then converted to their corresponding aryllactates by phenyllactate dehydrogenase (FldH, CLOSPO_00316). The aryllactates are then dehydrated by phenyllactate dehydratase (FldBC, CLOSPO_00310-311) along with its activator (FldI, CLOSPO_00309). Previous studies indicate that the dehydration reaction requires the substrate to first be activated to a CoA thioester, probably catalysed by acyl-CoA ligase (FldL, CLOSPO_00307), and that the CoA is recycled by the action of acyl-CoA transferase (FldA, CLOSPO_00308). Finally, the arylacrylates are reduced by acyl-CoA dehydrogenase (AcdA, CLOSPO_00312) involving its two electron transport factors (EtfA–EtfB, CLOSPO_00313–314). For the oxidative pathway, phenylpyruvate and 4-OH-phenylpyruvate are first oxidatively decarboxylated by pyruvate:ferredoxin oxidoreductase A (PorA, CLOSPO_00147-149), followed by phosphate acyltransferase and acyl kinase reactions that remain to be studied. The enzyme involved in indoleacetic acid production remains unknown, however, candidate genes in the genome include pyruvate:ferredoxin oxidoreductases B and C (PorB, CLOSPO_02262; PorC, CLOSPO_03792). Transformations for which the specific genes involved are not known are indicated in red.
Extended Data Figure 4 acdA, fldH and porA mutants exhibit growth defects when cultured with amino acids as the sole carbon source.
a–f, The wild-type (a–f) and acdA (a, d), fldH (b, e) and porA (c, f) mutant strains of C. sporogenes were cultured in minimal defined medium containing 10 amino acids at standard concentrations (SACC) with (d–f) or without (a–c) glucose (10 mM) and growth was monitored spectrophotometrically. Representative curves from n = 3 biological replicates are shown and doubling times are reported as mean ± s.d.
Extended Data Figure 5 fldZ does not exhibit a growth defect during amino acid fermentation.
a, b, The wild-type and fldZ mutant strains of C. sporogenes were cultured in minimal defined medium containing 10 amino acids at standard concentrations (SACC) with (a) or without (b) glucose (10 mM) and growth was monitored spectrophotometrically. Representative curves from n = 3 biological replicates are shown and doubling times are reported as mean ± s.d.
Extended Data Figure 6 The fldZ mutant is defective in conversion of phenylacrylate and 4-OH-phenylacryalte.
Resting cell suspensions of wild-type and fldZ mutant C. sporogenes were prepared and incubated with phenylacrylate, 4-OH-phenylacrylate or indoleacrylate for 1 h at 37 °C. Cells were then collected by centrifugation and the corresponding arylacrylates were detected in the supernatant by LC–MS/MS. The presence of a peak corresponds to a defect in the conversion of the indicated substrate (that is, retention of precursor). Representative chromatograms from n = 3 biological replicates are shown.
Extended Data Figure 7 porA is important for phenylacetate and 4-OH-phenylacetate production, but not indoleacetate.
Resting cell suspensions of wild-type and porA mutant C. sporogenes were prepared at a fourfold higher cell density (for example, 1/20 culture volume), and incubated with phenylalanine, tyrosine, tryptophan (red) or indolepyruvate (green) for 1 h at 37 °C. Cells were then collected by centrifugation and the arylacetates were detected in the supernatant by LC–MS/MS. The absence of a peak indicates a defect in the conversion of the indicated substrate to the corresponding arylacetate. Representative chromatograms from n = 3 biological replicates are shown.
Extended Data Figure 8 Broad overview of scaffold maps of mass cytometry data.
Scaffold maps of immune cells in the peripheral blood of mice colonized with wild-type or fldC mutant C. sporogenes. The full scaffold maps are presented here to provide topological context; zoomed-in regions are shown in Extended Data Fig. 9.
Extended Data Figure 9 Fine detail scaffold maps of mass cytometry data.
Scaffold maps of mass cytometry data from the peripheral blood of mice colonized with wild-type (top) or fldC mutant (bottom) C. sporogenes. Red nodes represent landmarks, or canonical immune cell populations defined manually. These landmarks facilitate the interpretation of the graph. Other nodes represent unsupervised clusters of similar cells, providing a data-driven representation of the cells present in the samples. The size of these nodes reflects the number of cells in that particular cluster. Unsupervised clusters are coloured according to their nearest landmark node. Edges in the graphs connect similar cells, with the length of each edge inversely proportional to that similarity. Cells that are most similar to one another are thereby connected by a short edge.
Extended Data Figure 10 Unsupervised force-directed graph of T cells in the peripheral blood.
T cells from mice colonized with wild-type and fldC mutant C. sporogenes were clustered together. Each cluster of T cells is coloured by the log2 fold change of the frequency in mice colonized with fldC mutant over wild-type C. sporogenes to visualize differences between the groups.
Supplementary information
Supplementary Information
This file contains supplementary tables 1-8. (PDF 483 kb)
Rights and permissions
About this article
Cite this article
Dodd, D., Spitzer, M., Van Treuren, W. et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652 (2017). https://doi.org/10.1038/nature24661
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24661
This article is cited by
-
The gut metabolite indole-3-propionic acid activates ERK1 to restore social function and hippocampal inhibitory synaptic transmission in a 16p11.2 microdeletion mouse model
Microbiome (2024)
-
Microbiota-derived indoles alleviate intestinal inflammation and modulate microbiome by microbial cross-feeding
Microbiome (2024)
-
Effects of dietary fibre on metabolic health and obesity
Nature Reviews Gastroenterology & Hepatology (2024)
-
Gut microbial metabolite facilitates colorectal cancer development via ferroptosis inhibition
Nature Cell Biology (2024)
-
Microbiota–gut–brain axis and its therapeutic applications in neurodegenerative diseases
Signal Transduction and Targeted Therapy (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.