Abstract
Gut bacteria face a key problem in how they capture enough energy to sustain their growth and physiology. The gut bacterium Clostridium sporogenes obtains its energy by utilizing amino acids in pairs, coupling the oxidation of one to the reduction of another—the Stickland reaction. Oxidative pathways produce ATP via substrate-level phosphorylation, whereas reductive pathways are thought to balance redox. In the present study, we investigated whether these reductive pathways are also linked to energy generation and the production of microbial metabolites that may circulate and impact host physiology. Using metabolomics, we find that, during growth in vitro, C. sporogenes produces 15 metabolites, 13 of which are present in the gut of C. sporogenes-colonized mice. Four of these compounds are reductive Stickland metabolites that circulate in the blood of gnotobiotic mice and are also detected in plasma from healthy humans. Gene clusters for reductive Stickland pathways suggest involvement of electron transfer proteins, and experiments in vitro demonstrate that reductive metabolism is coupled to ATP formation and not just redox balance. Genetic analysis points to the broadly conserved Rnf complex as a key coupling site for energy transduction. Rnf complex mutants show aberrant amino acid metabolism in a defined medium and are attenuated for growth in the mouse gut, demonstrating a role of the Rnf complex in Stickland metabolism and gut colonization. Our findings reveal that the production of circulating metabolites by a commensal bacterium within the host gut is linked to an ATP-yielding redox process.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The authors declare that the data supporting the findings of the present study are available within the paper and the Supplementary Information. Genome sequences analysed as part of the NIH Human Microbiome Project are available on NCBI GenBank under BioProject accession no. 43021.
Code availability
No customized code was used in the present study.
References
Van Treuren, W. & Dodd, D. Microbial contribution to the human metabolome: implications for health and disease. Annu. Rev. Pathol. 15, 345–369 (2020).
Russell, W. R. et al. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 57, 523–535 (2013).
Smith, E. A. & Macfarlane, G. T. Dissimilatory amino acid metabolism in human colonic bacteria. Anaerobe 3, 327–337 (1997).
Liu, Y., Hou, Y., Wang, G., Zheng, X. & Hao, H. Gut microbial metabolites of aromatic amino acids as signals in host–microbe interplay. Trends Endocrinol. Metab. 31, 818–834 (2020).
Allison, M. J., Bryant, M. P. & Doetsch, R. N. Volatile fatty acid growth factor for cellulolytic cocci of bovine rumen. Science 128, 474–475 (1958).
Stack, R. J., Hungate, R. E. & Opsahl, W. P. Phenylacetic acid stimulation of cellulose digestion by Ruminococcus albus 8. Appl. Environ. Microbiol. 46, 539–544 (1983).
Hungate, R. E. & Stack, R. J. Phenylpropanoic acid: growth factor for Ruminococcus albus. Appl. Environ. Microbiol. 44, 79–83 (1982).
Stickland, L. H. Studies in the metabolism of the strict anaerobes (genus Clostridium): the chemical reactions by which Cl. sporogenes obtains its energy. Biochem. J. 28, 1746–1759 (1934).
Nisman, B. The Stickland reaction. Bacteriol. Rev. 18, 16–42 (1954).
Lovitt, R. W., Kell, D. B. & Morris, J. G. Proline reduction by Clostridium sporogenes is coupled to vectorial proton ejection. FEMS Microbiol. Lett. 36, 269–273 (1986).
Bader, J. & Simon, H. ATP formation is coupled to the hydrogenation of 2-enoates in Clostridium sporogenes. FEMS Microbiol. Lett. 20, 171–175 (1983).
Dickert, S., Pierik, A. J. & Buckel, W. Molecular characterization of phenyllactate dehydratase and its initiator from Clostridium sporogenes. Mol. Microbiol. 44, 49–60 (2002).
Buckel, W. & Thauer, R. K. Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem. Rev. 118, 3862–3886 (2018).
Kimura, R. & Liao, T. H. A new thiamine decomposing anaerobic bacterium, Clostridium thiaminolyticum Kimura et Liao. Proc. Jpn Acad. 29, 132–133 (1953).
Karu, N. et al. A review on human fecal metabolomics: methods, applications and the human fecal metabolome database. Anal. Chim. Acta 1030, 1–24 (2018).
Wildenauer, F. X. & Winter, J. Fermentation of isoleucine and arginine by pure and syntrophic cultures of Clostridium sporogenes. FEMS Microbiol. Lett. 38, 373–379 (1986).
Lovitt, R. W., Morris, J. G. & Kell, D. B. The growth and nutrition of Clostridium sporogenes NCIB 8053 in defined media. J. Appl. Bacteriol. 62, 71–80 (1987).
Levin, B. J. et al. A prominent glycyl radical enzyme in human gut microbiomes metabolizes trans-4-hydroxy-l-proline. Science https://doi.org/10.1126/science.aai8386 (2017).
Lovitt, R. W., Kell, D. B. & Morris, J. G. The physiology of Clostridium sporogenes NCIB 8053 growing in defined media. J. Appl. Bacteriol. 62, 81–92 (1987).
Neumann-Schaal, M., Hofmann, J. D., Will, S. E. & Schomburg, D. Time-resolved amino acid uptake of Clostridium difficile 630(delta)erm and concomitant fermentation product and toxin formation. BMC Microbiol. 15, 281 (2015).
Bouillaut, L., Self, W. T. & Sonenshein, A. L. Proline-dependent regulation of Clostridium difficile Stickland metabolism. J. Bacteriol. 195, 844–854 (2013).
Jackson, S., Calos, M., Myers, A. & Self, W. T. Analysis of proline reduction in the nosocomial pathogen Clostridium difficile. J. Bacteriol. 188, 8487–8895 (2006).
Ragsdale, S. W. Pyruvate ferredoxin oxidoreductase and its radical intermediate. Chem. Rev. 103, 2333–2346 (2003).
Xu, X. L. & Grant, G. A. Identification and characterization of two new types of bacterial l-serine dehydratases and assessment of the function of the ACT domain. Arch. Biochem. Biophys. 540, 62–69 (2013).
Leach, S., Harvey, P. & Wali, R. Changes with growth rate in the membrane lipid composition of and amino acid utilization by continuous cultures of Campylobacter jejuni. J. Appl. Microbiol. 82, 631–640 (1997).
Velayudhan, J., Jones, M. A., Barrow, P. A. & Kelly, D. J. l-Serine catabolism via an oxygen-labile l-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect. Immun. 72, 260–268 (2004).
Claus, S. P. et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 4, 219 (2008).
Dodd, D. et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652 (2017).
Guo, C. J. et al. Depletion of microbiome-derived molecules in the host using Clostridium genetics. Science https://doi.org/10.1126/science.aav1282 (2019).
Sharon, G. et al. Human gut microbiota from autism spectrum disorder promote behavioral symptoms in mice. Cell 177, 1600–1618.e1617 (2019).
Aronov, P. A. et al. Colonic contribution to uremic solutes. J. Am. Soc. Nephrol. 22, 1769–1776 (2011).
Fonknechten, N. et al. Clostridium sticklandii, a specialist in amino acid degradation:revisiting its metabolism through its genome sequence. BMC Genom. 11, 555 (2010).
Stadtman, T. C. & Elliott, P. Studies on the enzymic reduction of amino acids. II. Purification and properties of d-proline reductase and a proline racemase from Clostridium sticklandii. J. Biol. Chem. 228, 983–997 (1957).
Barker, H. A. Amino acid degradation by anaerobic bacteria. Annu. Rev. Biochem. 50, 23–40 (1981).
Nemet, I. et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell 180, 862–877.e822 (2020).
Seedorf, H. et al. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc. Natl Acad.Sci. USA 105, 2128–2133 (2008).
Li, F. et al. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J. Bacteriol. 190, 843–850 (2008).
Herrmann, G., Jayamani, E., Mai, G. & Buckel, W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190, 784–791 (2008).
Kuhns, M. et al. The Rnf complex from the acetogenic bacterium Acetobacterium woodii: purification and characterization of RnfC and RnfB. Biochim. Biophys. Acta Bioenerg. 1861, 148263 (2020).
Hreha, T. N. et al. Complete topology of the RNF complex from Vibrio cholerae. Biochemistry 54, 2443–2455 (2015).
Nayfach, S., Fischbach, M. A. & Pollard, K. S. MetaQuery: a web server for rapid annotation and quantitative analysis of specific genes in the human gut microbiome. Bioinformatics 31, 3368–3370 (2015).
Steed, A. L. et al. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357, 498–502 (2017).
Venkatesh, M. et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41, 296–310 (2014).
Medema, M. H., Takano, E. & Breitling, R. Detecting sequence homology at the gene cluster level with MultiGeneBlast. Mol. Biol. Evol. 30, 1218–1223 (2013).
Acknowledgements
We thank J. Sonnenburg, M. Fischbach, C. Walsh, A. Southwick, C. Fischer and I. Cann for valuable discussions. We thank M. Wu for assistance with amino acid analysis by LC–MS, M. St. Onge with help constructing mutants, C.-J. Guo and R. De La Pena for assistance with GC–MS analysis of short-chain fatty acids and A. Dimas and D. Nguyen for assistance with gnotobiotic animal experiments. This work was funded in part by NIH grants (nos. K08-DK110335 and R35-GM142873 to D.D.).
Author information
Authors and Affiliations
Contributions
Y.L. and D.D. conceived the project, designed and performed experiments, analysed data, wrote the original draft and revised it with comments from all the authors. H.C. assisted with MS method development and data analysis. W.V.T. and B.H. contributed data. S.K.H. assisted with animal experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Sam Light and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 C. sporogenes pathways converge on proline reductase.
Arginine, citrulline, ornithine, and trans-4-hydroxy-L-proline pathways all converge on L-proline which is converted to D-proline by proline racemase. D-proline is then converted to 5-aminovalerate by the enzyme, proline reductase. Locus tag IDs from the C. sporogenes ATCC 15579 genome are provided next to each of the enzyme-catalyzed steps. For the proline reductase enzyme, the locus tag IDs are provided for each of the two gene clusters which contain multiple copies of proline reductase enzyme subunits.
Extended Data Fig. 2 Proline reductase gene clusters and in vitro phenotypes of Clostridium sporogenes mutants.
A) The two gene clusters for proline metabolism in C. sporogenes encode multiple copies of the prdA, prdB, prdC, and prdG genes. Red arrows designate mutants tested in this study for proline metabolism. Locus tag IDs for the two gene clusters are provided. B) Wild-type C. sporogenes and two ClosTron insertional mutants in prdR were cultured in defined medium containing 20 amino acids and 5-aminovalerate in the supernatant was quantified at 3, 6, 24, and 48 h by LC-MS. Numbers after the prdR name indicate positions in the gene where the group II intron was integrated. Experiments were performed in triplicate and data are reported as means ± standard deviations. C) Wild-type C. sporogenes and a prdF mutant were cultured in defined medium containing 20 amino acids and arginine and proline in the supernatant was quantified at 3, 6, 24, and 48 h by LC-MS. Data are plotted as means ± standard deviations from n = 3 experiments.
Extended Data Fig. 3 Stickland metabolites do not arise from de novo biosynthetic pathways.
A-D) Stable isotope tracing. C. sporogenes was cultured in a synthetic medium containing 20 amino acids where Phe, Tyr, Trp, and Pro were individually substituted by their deuterium isotopologues. Cell-free supernatants were collected at t = 24 h and metabolites were detected by LC-MS. Data are plotted as means ± standard deviations from n = 3 cultures. A-C) For phenylpropionate, 3-(4-hydroxyphenyl)propionate, and indolepropionate, no unlabeled products were detected when isotopically labeled amino acids were provided. D) Unlabeled 5-aminovalerate levels were reduced when isotopically labeled Pro was supplied, but ~1 mM 5-aminovalerate was still detected suggesting another source exists for its production. E) Stable isotope tracing of C. sporogenes cell suspensions incubated with stable isotopically labeled arginine (Arg-d7). When cells were incubated with Arg-d7, labeled proline and 5-aminovalerate were detected, suggesting that arginine is a substrate for 5-aminovalerate production. Data are plotted as means ± standard deviations from n = 3 experiments.
Extended Data Fig. 4 Indolepropionate (IPA) and phenylpropionate (PPA) production is stimulated by incubation with oxidative Stickland amino acids.
A) C. sporogenes cell suspensions were incubated with stable isotopically labeled tryptophan (Trp-d5) alone or in combination with oxidative Stickland amino acids (Ile, Leu, Val, or Met), then labeled IPA-d5 was measured by LC-MS. IPA-d5 levels were stimulated by addition of Leu, Val, or Met. B) C. sporogenes cell suspensions were incubated with stable isotopically labeled phenylalanine (Phe-d8) alone or in combination with oxidative Stickland amino acids (Ile, Leu, Val, or Met), then labeled PPA-d6 was measured by LC-MS. PPA-d6 levels were stimulated by addition of Leu, Val, or Met. Data are plotted as means ± standard deviations from n = 3 experiments.
Extended Data Fig. 5 Cinnamate reduction is coupled to ATP formation in C. sporogenes.
A) The cinnamate reductase gene encodes a unique FAD/[FeS]/FMN containing enzyme with separate binding sites for NADH, cinnamate, and the artificial electron carrier methylviologen, however the natural electron carrier remains unknown. B) Resting cell suspensions of C. sporogenes accumulate ATP after being incubated with cinnamate. Buffer control is shown. For B, experiments were repeated independently three times and representative data are shown.
Extended Data Fig. 6 Reductive metabolism of D-phenyllactate is coupled to ATP formation involving acdA.
A) D- or L-phenyllactate was added (1 mM) to resting cell suspensions of C. sporogenes, and ATP levels were measured at indicated time points using a luciferase-based assay. B) Reductive (PPA) and oxidative (PAA) pathway end products were measured by LC-MS during D-phenyllactate metabolism. PPA, phenylpropionate; PAA, phenylacetate. C) Pathways for oxidative and reductive metabolism of phenylalanine showing the position of AcdA in the pathway. D-E) Phenylacetate (D) or phenylpropionate (E) was added (1 mM) to resting cell suspensions of C. sporogenes, and ATP levels were measured at indicated time points using a luciferase-based assay. F-H) DL-phenyllactate (F), DL-3-(4-hydroxyphenyl)lactate (G), or DL-indolelactate (H) was added (1 mM) to resting cell suspensions of WT or acdA mutant C. sporogenes, and ATP levels were measured at indicated time points using a luciferase-based assay. For A-B, D-H, experiments were repeated independently three times and representative data are shown.
Extended Data Fig. 7 The protonophore, 3,3′,4′,5-tetrachlorosalicylanilide (TCS), uncouples reductive Stickland metabolism from ATP formation.
Resting cell suspensions of C. sporogenes were preincubated with ethanol (vehicle) or varying concentrations of TCS for 30 min. Then substrates (DL-phenyllactate (DL-PLA) or proline (Pro)) were added, and aliquots were taken at different time-points and quenched in DMSO. Total cellular ATP was quantified using a luciferase-based assay and normalized to total cellular protein and substrates/metabolites were quantified using LC-MS. A) Dose dependent decrease in ATP formation from DL-phenyllactate with increasing concentrations of TCS. B) Conversion of DL-phenyllactate to phenylpropionate (PPA) at the 15 min timepoint remains constant with increasing TCS while (C) ATP production diminishes. D) Conversion of DL-phenyllactate to phenylpropionate (PPA) increases over time irrespective of TCS levels. E) Dose dependent decrease in ATP formation from Pro with increasing concentrations of TCS. F) Conversion of Pro to 5-aminovalerate (5-AVA) at the 15 min timepoint decreases but is not completely blocked with increasing TCS while (G) ATP production diminishes becoming negligible at 100 μM TCS. H) Conversion of Pro to 5-aminovalerate (5-AVA) increases over time, and while the rate of conversion decreases with increasing TCS levels, proline conversion to 5-aminovalerate is complete by the 60 min timepoint.
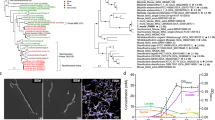
Extended Data Fig. 8 The Rnf gene cluster is widely distributed among reference genomes from the Human Microbiome Project.
A) 16 S rRNA tree of organisms from the human microbiome reference genome collection determined to have Rnf gene clusters as determined by BLASTp and manual inspection of gene neighborhoods using MultiGeneBlast. B) Rnf gene clusters for select organisms (corresponding to triangles in panel A), grouped by phyla.
Extended Data Fig. 9 The enoate is a chemical moiety common to pathways for microbial metabolites.
Enoates or their enoyl-CoA derivatives are common intermediates that serve as alternate electron acceptors for anaerobic respiration. Enzymes catalyzing the reduction of enoates or enoyl-CoAs are shown.
Extended Data Fig. 10 Revised model for C. sporogenes Stickland metabolism and its contribution to gut bacterial derived metabolites in the gut.
Oxidative pathways yield ATP directly via substrate level phosphorylation, whereas reductive pathways generate reduced ferredoxin via flavin-based electron bifurcation contributing to a proton (or sodium ion) motive force via the Rnf complex. Protons or sodium ions translocated by the Rnf complex re-enter the cell via the membrane bound ATP synthase, resulting in the synthesis of ATP. The membrane gradient may also contribute to other physiological processes such as membrane transport and chemotaxis (not shown).
Supplementary information
Supplementary Information
Supplementary Figs. 1–4.
Supplementary Tables
Supplementary Tables 1–27.
Rights and permissions
About this article
Cite this article
Liu, Y., Chen, H., Van Treuren, W. et al. Clostridium sporogenes uses reductive Stickland metabolism in the gut to generate ATP and produce circulating metabolites. Nat Microbiol 7, 695–706 (2022). https://doi.org/10.1038/s41564-022-01109-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-022-01109-9
This article is cited by
-
gutSMASH predicts specialized primary metabolic pathways from the human gut microbiota
Nature Biotechnology (2023)
-
Host-microbe co-metabolism via MCAD generates circulating metabolites including hippuric acid
Nature Communications (2023)
-
Rice flowering improves the muscle nutrient, intestinal microbiota diversity, and liver metabolism profiles of tilapia (Oreochromis niloticus) in rice-fish symbiosis
Microbiome (2022)
-
Stickland metabolism in the gut
Nature Microbiology (2022)