Abstract
The most prevalent route of HIV-1 infection is across mucosal tissues after sexual contact. Langerhans cells (LCs) belong to the subset of dendritic cells (DCs) that line the mucosal epithelia of vagina and foreskin and have the ability to sense and induce immunity to invading pathogens1. Anatomical and functional characteristics make LCs one of the primary targets of HIV-1 infection2. Notably, LCs form a protective barrier against HIV-1 infection and transmission3,4,5. LCs restrict HIV-1 infection through the capture of HIV-1 by the C-type lectin receptor Langerin and subsequent internalization into Birbeck granules5. However, the underlying molecular mechanism of HIV-1 restriction in LCs remains unknown. Here we show that human E3-ubiquitin ligase tri-partite-containing motif 5α (TRIM5α) potently restricts HIV-1 infection of LCs but not of subepithelial DC-SIGN+ DCs. HIV-1 restriction by TRIM5α was thus far considered to be reserved to non-human primate TRIM5α orthologues6,7,8,9, but our data strongly suggest that human TRIM5α is a cell-specific restriction factor dependent on C-type lectin receptor function. Our findings highlight the importance of HIV-1 binding to Langerin for the routeing of HIV-1 into the human TRIM5α-mediated restriction pathway. TRIM5α mediates the assembly of an autophagy-activating scaffold to Langerin, which targets HIV-1 for autophagic degradation and prevents infection of LCs. By contrast, HIV-1 binding to DC-SIGN+ DCs leads to disassociation of TRIM5α from DC-SIGN, which abrogates TRIM5α restriction. Thus, our data strongly suggest that restriction by human TRIM5α is controlled by C-type-lectin-receptor-dependent uptake of HIV-1, dictating protection or infection of human DC subsets. Therapeutic interventions that incorporate C-type lectin receptors and autophagy-targeting strategies could thus provide cell-mediated resistance to HIV-1 in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Banchereau, J. et al. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18, 767–811 (2000)
Hladik, F. et al. Initial events in establishing vaginal entry and infection by human immunodeficiency virus type-1. Immunity 26, 257–270 (2007)
Ribeiro, C. M. S., Sarrami-Forooshani, R. & Geijtenbeek, T. B. H. HIV-1 border patrols: Langerhans cells control antiviral responses and viral transmission. Future Virol. 10, 1231–1243 (2015)
Sarrami-Forooshani, R. et al. Human immature Langerhans cells restrict CXCR4-using HIV-1 transmission. Retrovirology 11, 52 (2014)
de Witte, L. et al. Langerin is a natural barrier to HIV-1 transmission by Langerhans cells. Nat. Med. 13, 367–371 (2007)
Stremlau, M. et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 (2004)
Sawyer, S. L., Wu, L. I., Emerman, M. & Malik, H. S. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl Acad. Sci. USA 102, 2832–2837 (2005)
Song, B. et al. Retrovirus restriction by TRIM5α variants from Old World and New World primates. J. Virol. 79, 3930–3937 (2005)
Sayah, D. M., Sokolskaja, E., Berthoux, L. & Luban, J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430, 569–573 (2004)
van den Berg, L. M. et al. Caveolin-1 mediated uptake via Langerin restricts HIV-1 infection in human Langerhans cells. Retrovirology 11, 123 (2014)
Stremlau, M. et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl Acad. Sci. USA 103, 5514–5519 (2006)
Yap, M. W., Nisole, S. & Stoye, J. P. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr. Biol. 15, 73–78 (2005)
Ganser-Pornillos, B. K. et al. Hexagonal assembly of a restricting TRIM5α protein. Proc. Natl Acad. Sci. USA 108, 534–539 (2011)
de Jong, M. A. et al. Mutz-3-derived Langerhans cells are a model to study HIV-1 transmission and potential inhibitors. J. Leukoc. Biol. 87, 637–643 (2010)
Mandell, M. A. et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell 30, 394–409 (2014)
Fujita, N. et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J. Cell Biol. 203, 115–128 (2013)
Moreau, K., Ravikumar, B., Renna, M., Puri, C. & Rubinsztein, D. C. Autophagosome precursor maturation requires homotypic fusion. Cell 146, 303–317 (2011)
Gramberg, T. et al. Interactions of LSECtin and DC-SIGN/DC-SIGNR with viral ligands: differential pH dependence, internalization and virion binding. Virology 373, 189–201 (2008)
Geijtenbeek, T. B. et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100, 587–597 (2000)
Gringhuis, S. I. et al. HIV-1 exploits innate signaling by TLR8 and DC-SIGN for productive infection of dendritic cells. Nat. Immunol. 11, 419–426 (2010)
Blanchet, F. P. et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity 32, 654–669 (2010)
Ward, E. M., Stambach, N. S., Drickamer, K. & Taylor, M. E. Polymorphisms in human Langerin affect stability and sugar binding activity. J. Biol. Chem. 281, 15450–15456 (2006)
Gringhuis, S. I., den Dunnen, J., Litjens, M., van der Vlist, M. & Geijtenbeek, T. B. Carbohydrate-specific signaling through the DC-SIGN signalosome tailors immunity to Mycobacterium tuberculosis, HIV-1 and Helicobacter pylori . Nat. Immunol. 10, 1081–1088 (2009)
Smith, A. L. et al. Leukocyte-specific protein 1 interacts with DC-SIGN and mediates transport of HIV to the proteasome in dendritic cells. J. Exp. Med. 204, 421–430 (2007)
Wu, X., Anderson, J. L., Campbell, E. M., Joseph, A. M. & Hope, T. J. Proteasome inhibitors uncouple rhesus TRIM5α restriction of HIV-1 reverse transcription and infection. Proc. Natl Acad. Sci. USA 103, 7465–7470 (2006)
Perez-Caballero, D., Hatziioannou, T., Yang, A., Cowan, S. & Bieniasz, P. D. Human tripartite motif 5α domains responsible for retrovirus restriction activity and specificity. J. Virol. 79, 8969–8978 (2005)
Stremlau, M., Perron, M., Welikala, S. & Sodroski, J. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79, 3139–3145 (2005)
Song, B. et al. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol. 79, 6111–6121 (2005)
Shi, J. & Aiken, C. Saturation of TRIM5α-mediated restriction of HIV-1 infection depends on the stability of the incoming viral capsid. Virology 350, 493–500 (2006)
Kootstra, N. A., Munk, C., Tonnu, N., Landau, N. R. & Verma, I. M. Abrogation of postentry restriction of HIV-1-based lentiviral vector transduction in simian cells. Proc. Natl Acad. Sci. USA 100, 1298–1303 (2003)
Arrighi, J. F. et al. DC-SIGN-mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. J. Exp. Med. 200, 1279–1288 (2004)
Björndal, A. et al. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71, 7478–7487 (1997)
Setiawan, L. C. & Kootstra, N. A. Adaptation of HIV-1 to rhTrim5α-mediated restriction in vitro . Virology 486, 239–247 (2015)
Eng, K. E., Panas, M. D., Karlsson Hedestam, G. B. & McInerney, G. M. A novel quantitative flow cytometry-based assay for autophagy. Autophagy 6, 634–641 (2010)
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 (2012)
Chan, L. L. et al. A novel image-based cytometry method for autophagy detection in living cells. Autophagy 8, 1371–1382 (2012)
Acknowledgements
We are grateful to the members of the Host Defense group and Laboratory for Viral Immune Pathogenesis (Department of Experimental Immunology, Academic Medical Center, Amsterdam, The Netherlands) for their input and D. Picavet (van Leeuwenhoek Centrum for Advanced Microscopy, Academic Medical Center, Amsterdam, The Netherlands) for technical assistance during confocal experiments. We wish to thank the Boerhaave Medical Centre (Amsterdam, The Netherlands) and A. Knottenbelt (Flevoclinic, Almere, The Netherlands) for the provision of human skin tissues. This work was supported by the Dutch Scientific Organization NWO (VENI 863.13.025 and VICI 918.10.619), Aids Fonds (2010038) and European Research Council (Advanced grant 670424).
Author information
Authors and Affiliations
Contributions
C.M.S.R. designed, performed and interpreted most experiments and prepared the manuscript; R.S.F. assisted with the lentiviral transductions and the confocal experiments; L.C.S. assisted with culturing the CD4+CCR5+ U87 cell lines and with the lentiviral transductions; E.M.Z.W. cultured MUTZ-LCs and helped with immunoblotting; J.L.v.H. assisted with primary cell isolation and silencing experiments; W.T. and N.N.v.d.W. performed the EM microscopy; N.A.K. and S.I.G. helped prepare the manuscript and T.B.H.G. supervised all aspects of the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks J. Luban, C. Munz and G. Towers for their contribution to the peer review of this work.
Extended data figures and tables
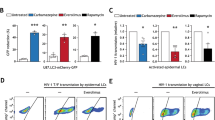
Extended Data Figure 1 Langerin in MUTZ-LCs restricts HIV-1 integration, infection and transmission to CD4+ T cells.
a, b, HIV-1NL4.3 integration (a) and infection (b) of MUTZ-LCs after Langerin silencing, determined by Alu-PCR (a) and intracellular p24 staining (b). c, HIV-1NL4.3-BaL transmission by MUTZ-LCs after Langerin silencing, determined in LC and T-cell coculture by intracellular p24 staining. d, e, Silencing was confirmed by real-time PCR (d) or by flow cytometer (e; representative of n = 3). mRNA expression was normalized to β-actin (d) and set at 1 in control-siRNA treated cells. *P < 0.05 (two-tailed t-test). Data are mean ± s.d. of three (a, c, d) and four (b) independent experiments.
Extended Data Figure 2 Silencing of TRIM5α, Atg5, Atg16L1 and LSP-1 by RNA interference.
a–k, Indicated proteins were silenced using specific SMARTpools and non-targeting siRNA as a control. Silencing was confirmed by real-time PCR (a–g) or by immunoblotting (β-actin served as loading control; h–k) in MUTZ-LCs (a, d, e, h, i, j), primary LCs (b), DCs (c), CD4+CCR5+ U87 parental cells (f) or CD4+CCR5+ U87 cells transduced with either Langerin (f, k) or rhesus TRIM5α (g). mRNA expression was normalized to β-actin (a, d, e) or GAPDH (b, c, f, g) and set at 1 in cells treated with control siRNA. Relative abundance of indicated proteins was quantified by normalizing to β-actin and set at 1 in control siRNA treated cells. Representative of n = 2 (d, e, h–k). For gel source data, see Supplementary Fig. 1. Data are mean ± s.d. of three (a, c, f, g) and six (b) independent experiments.
Extended Data Figure 3 Human TRIM5α-mediated restriction in LCs or, the lack thereof in DCs, is independent of virus tropism.
a–d, HIV-1NL4.3 (X4, CXCR4-tropic virus) or HIV-1NL4.3-BaL (R5, CCR5-tropic virus) integration (a, c) and infection (b, d) of primary LCs (a, b) or DCs (b, d) after TRIM5α silencing determined by Alu-PCR (a, c) and intracellular p24 staining (b, d). *P < 0.05, **P < 0.01 (two-tailed t-test). Data are mean ± s.d. of three (a–d) independent experiments.
Extended Data Figure 4 ULK1 complex-dependent autophagy restricts HIV-1 integration in LCs and human TRIM5α restriction is dependent on Atg5 function.
a, TRIM5α, p62 and Atg16L1 in whole-cell lysates of uninfected MUTZ-LCs before (input) or after immunoprecipitation with Atg16L1, p62, TRIM5α, rabbit IgG control (as control for Atg16L1 and TRIM5α IP) or mouse IgG2a isotype control (as control for p62 immunoprecipitation), determined by immunoblotting (n.d., not determined). b, Autophagy induction in primary LCs pre-treated with bafilomycin followed by incubation with HIV-1NL4.3, determined by immunoblotting for LC3. For gel source data, see Supplementary Fig. 1. c, HIV-1NL4.3 infection of MUTZ-LCs after Atg5 or Atg16L1 silencing, determined by intracellular p24 staining. d, HIV-1NL4.3 integration into MUTZ-LCs after Atg13 or FIP200 silencing, determined by Alu-PCR. e, f, HIV-1NL4.3 integration (e) or infection (f) of MUTZ-LCs after Atg5, TRIM5α silencing or simultaneously with Atg5 and TRIM5α silencing, determined by Alu-PCR (e) and intracellular p24 staining (f). Data are representative of three (a) or two (b, d–f) experiments and mean ± s.d. of four independent experiments (c).
Extended Data Figure 5 Increased Atg5 recruitment into TRIM5α–Atg16L1 complex scaffold in CD4+CCR5+ U87 transfectants.
a, Atg5, TRIM5α and Atg16L1 in whole-cell lysates of CD4+CCR5+ U87 parental cells (U87) or transduced with either human TRIM5α (U87 hu5α) or rhesus TRIM5α (U87 rh5α) infected with HIV-1NL4.3-BaL before (input) or after immunoprecipitation with Atg16L1 or rabbit IgG control, determined by immunoblotting (n.d., not determined). b, Autophagy induction in U87 transfectants with bafilomycin followed by incubation with HIV-1SF162, determined by immunoblotting for LC3 (autophagy induction in control CD4+CCR5+ U87 parental cells presented in Fig. 3o). Relative abundance of LC3 II determined by normalizing to β-actin. Representative of n = 2 (a, b). For gel source data, see Supplementary Fig. 1. c, HIV-1SF162 infection of CD4+CD5+ U87 cells transduced with rhesus TRIM5α after Atg16L1 silencing, determined by intracellular p24 staining. *P < 0.05 (t-test). Data are mean ± s.d. of three (c) independent experiments.
Extended Data Figure 6 Human TRIM5α induces autophagy upon HIV-1 exposure in Langerin+ U87 transfectant and interacts with Langerin through LSP-1, but not Atg16L1.
a, Atg5, TRIM5α and Atg16L1 in whole-cell lysates of CD4+CCR5+ U87 parental cells (U87) or transduced with Langerin (U87 Langerin) before (input) or after immunoprecipitation with Atg16L1 or rabbit IgG control, determined by immunoblotting (n.d., not determined). b, Autophagy levels in Langerin+ U87 transfectant after TRIM5α silencing, pre-treated with bafilomycin followed by incubation with HIV-1NL4.3-BaL, determined by intracellular LC3 II levels by flow cytometer. c, LSP-1 in whole-cell lysates of MUTZ-LCs infected with HIV-1NL4.3 before (input) or after immunoprecipitation with TRIM5α or rabbit IgG control. d, TRIM5α in whole-cell lysates of Langerin+ U87 transfectant after Atg16L1 silencing before (input) or after immunoprecipitation with Langerin, determined by immunoblotting. For gel source data, see Supplementary Fig. 1. e, Autophagy induction in Langerin+ U87 transfectant pre-treated with bafilomycin followed by incubation with VSV-G-pseudotyped HIV-1, determined by intracellular LC3 II levels. Data are representative of two experiments (a–e).
Extended Data Figure 7 Proteosome inhibition does not relieve Langerin-mediated restriction of HIV-1 reverse-transcription products nor infection.
a–d, R/gag proviral DNA levels (a, c) and HIV-1 infection (b, d) in CD4+CCR5+ U87 parental cells (U87) or cells transduced with either rhesus TRIM5α (U87 rh5α) or Langerin (U87 Lang) after pre-treatment with proteosome inhibitor MG-132 and infected with VSV-G-pseudotyped HIV-1 (a, b; VSV-G) or HIV-1NL4.3-BaL (c, d; HIV-1), determined by qPCR (a, c) and intracellular p24 staining (b, d). Data are representative of two experiments (a–d).
Extended Data Figure 8 Langerin-controlled human TRIM5α restriction mechanism in Langerhans cells.
a, HIV-1 binding to Langerin in Langerhans cells drives human TRIM5α-mediated restriction of viral integration, HIV-1 infection and HIV-1 transmission to CD4+ T cells. b, Langerin associates at steady-state with LSP-1–TRIM5α–Atg16L1 complex. Capture of HIV-1 by Langerin targets internalization of the incoming virus into Birbeck granules. Upon viral fusion, human TRIM5α mediates recruitment of Atg5 to TRIM5α–Atg16L1–HIV-1p24 capsid complex, which promotes lipidation of LC3 (LC3 II) and thereby elicit autophagosome formation. Vesicles containing Langerin–HIV-1 capsid complexes are subsequently targeted into autophagosomes for lysosomal degradation, which prevents infection of Langerhans cells.
Supplementary information
Supplementary Information
DESCRIPThis file contains Supplementary Figure 1, gel source data with size marker indications for Figure 2a; Figure 3e,g,o; Figure 4e,i; Extended Data Fig. 2h-k, Extended Data Fig. 4a,b, Extended Data Fig. 5a,b and Extended Data Fig. 6a,c,d. It also contains Supplementary Table 1, primary data of HIV-1 integration Alu-PCR assay and calculation of relative HIV-1 integration; see also Figure 1a.TION (PDF 2796 kb)
Rights and permissions
About this article
Cite this article
Ribeiro, C., Sarrami-Forooshani, R., Setiawan, L. et al. Receptor usage dictates HIV-1 restriction by human TRIM5α in dendritic cell subsets. Nature 540, 448–452 (2016). https://doi.org/10.1038/nature20567
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20567
This article is cited by
-
The effects of CypA on apoptosis: potential target for the treatment of diseases
Applied Microbiology and Biotechnology (2024)
-
TRIM5α recruits HDAC1 to p50 and Sp1 and promotes H3K9 deacetylation at the HIV-1 LTR
Nature Communications (2023)
-
Immune activation of vaginal human Langerhans cells increases susceptibility to HIV-1 infection
Scientific Reports (2023)
-
HIV transmitting mononuclear phagocytes; integrating the old and new
Mucosal Immunology (2022)
-
Autophagy-enhancing drugs limit mucosal HIV-1 acquisition and suppress viral replication ex vivo
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.