Abstract
Chronic viral infections are characterized by a state of CD8+ T-cell dysfunction that is associated with expression of the programmed cell death 1 (PD-1) inhibitory receptor1,2,3,4. A better understanding of the mechanisms that regulate CD8+ T-cell responses during chronic infection is required to improve immunotherapies that restore function in exhausted CD8+ T cells. Here we identify a population of virus-specific CD8+ T cells that proliferate after blockade of the PD-1 inhibitory pathway in mice chronically infected with lymphocytic choriomeningitis virus (LCMV). These LCMV-specific CD8+ T cells expressed the PD-1 inhibitory receptor, but also expressed several costimulatory molecules such as ICOS and CD28. This CD8+ T-cell subset was characterized by a unique gene signature that was related to that of CD4+ T follicular helper (TFH) cells, CD8+ T cell memory precursors and haematopoietic stem cell progenitors, but that was distinct from that of CD4+ TH1 cells and CD8+ terminal effectors. This CD8+ T-cell population was found only in lymphoid tissues and resided predominantly in the T-cell zones along with naive CD8+ T cells. These PD-1+CD8+ T cells resembled stem cells during chronic LCMV infection, undergoing self-renewal and also differentiating into the terminally exhausted CD8+ T cells that were present in both lymphoid and non-lymphoid tissues. The proliferative burst after PD-1 blockade came almost exclusively from this CD8+ T-cell subset. Notably, the transcription factor TCF1 had a cell-intrinsic and essential role in the generation of this CD8+ T-cell subset. These findings provide a better understanding of T-cell exhaustion and have implications in the optimization of PD-1-directed immunotherapy in chronic infections and cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
The microarray data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE84105.
References
Zajac, A. J. et al. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188, 2205–2213 (1998)
Gallimore, A. et al. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J. Exp. Med. 187, 1383–1393 (1998)
Sharma, P. & Allison, J. P. The future of immune checkpoint therapy. Science 348, 56–61 (2015)
Barber, D. L. et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 439, 682–687 (2006)
Blackburn, S. D., Shin, H., Freeman, G. J. & Wherry, E. J. Selective expansion of a subset of exhausted CD8 T cells by αPD-L1 blockade. Proc. Natl Acad. Sci. USA 105, 15016–15021 (2008)
Blackburn, S. D. et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 10, 29–37 (2009)
Jin, H. T. et al. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc. Natl Acad. Sci. USA 107, 14733–14738 (2010)
Paley, M. A. et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science 338, 1220–1225 (2012)
Quigley, M. F., Gonzalez, V. D., Granath, A., Andersson, J. & Sandberg, J. K. CXCR5+ CCR7− CD8 T cells are early effector memory cells that infiltrate tonsil B cell follicles. Eur. J. Immunol. 37, 3352–3362 (2007)
Kim, H. J., Verbinnen, B., Tang, X., Lu, L. & Cantor, H. Inhibition of follicular T-helper cells by CD8+ regulatory T cells is essential for self tolerance. Nature 467, 328–332 (2010)
Monney, L. et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 415, 536–541 (2002)
Matloubian, M., Kolhekar, S. R., Somasundaram, T. & Ahmed, R. Molecular determinants of macrophage tropism and viral persistence: importance of single amino acid changes in the polymerase and glycoprotein of lymphocytic choriomeningitis virus. J. Virol. 67, 7340–7349 (1993)
Takeda, K., Kaisho, T. & Akira, S. Toll-like receptors. Annu. Rev. Immunol. 21, 335–376 (2003)
Crotty, S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011)
Yang, C. Y. et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12, 1221–1229 (2011)
Pearce, E. L. et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature 460, 103–107 (2009)
Cui, G. et al. IL-7-induced glycerol transport and TAG synthesis promotes memory CD8+ T cell longevity. Cell 161, 750–761 (2015)
Reya, T. et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423, 409–414 (2003)
Gerner, M. Y., Kastenmuller, W., Ifrim, I., Kabat, J. & Germain, R. N. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376 (2012)
Jung, Y. W., Rutishauser, R. L., Joshi, N. S., Haberman, A. M. & Kaech, S. M. Differential localization of effector and memory CD8 T cell subsets in lymphoid organs during acute viral infection. J. Immunol. 185, 5315–5325 (2010)
Mueller, S. N. et al. Viral targeting of fibroblastic reticular cells contributes to immunosuppression and persistence during chronic infection. Proc. Natl Acad. Sci. USA 104, 15430–15435 (2007)
Anderson, K. G. et al. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protocols 9, 209–222 (2014)
Haynes, N. M. et al. Role of CXCR5 and CCR7 in follicular TH cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J. Immunol. 179, 5099–5108 (2007)
Shiow, L. R. et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 440, 540–544 (2006)
Yamazaki, C. et al. Critical roles of a dendritic cell subset expressing a chemokine receptor, XCR1. J. Immunol. 190, 6071–6082 (2013)
Xu, L. et al. The transcription factor TCF-1 initiates the differentiation of TFH cells during acute viral infection. Nat. Immunol. 16, 991–999 (2015)
Choi, Y. S. et al. LEF-1 and TCF-1 orchestrate TFH differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat. Immunol. 16, 980–990 (2015)
Wu, J. Q. et al. Tcf7 is an important regulator of the switch of self-renewal and differentiation in a multipotential hematopoietic cell line. PLoS Genet . 8, e1002565 (2012)
Matloubian, M., Concepcion, R. J. & Ahmed, R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J. Virol. 68, 8056–8063 (1994)
Wherry, E. J., Blattman, J. N., Murali-Krishna, K., van der Most, R. & Ahmed, R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77, 4911–4927 (2003)
Masopust, D. et al. Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207, 553–564 (2010)
Murali-Krishna, K. et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity 8, 177–187 (1998)
Johnston, R. J. et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325, 1006–1010 (2009)
Ivanova, N. B. et al. A stem cell molecular signature. Science 298, 601–604 (2002)
Joshi, N. S. et al. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295 (2007)
Miyazaki, M. et al. The E-Id protein axis modulates the activities of the PI3K-AKT-mTORC1-Hif1a and c-myc/p19Arf pathways to suppress innate variant TFH cell development, thymocyte expansion, and lymphomagenesis. Genes Dev. 29, 409–425 (2015)
Allen, C. D. et al. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol. 5, 943–952 (2004)
Acknowledgements
This work was supported by National Institutes of Health grants R01 AI30048 (R.A.), P01 AI056299 (R.A. and A.H.S.), R01 AI112579 (H.H.X.) and R01 AI121080 (H.-H.X.) and also by the Intramural Research Program of NIAID, NIH (R.N.G. and M.Y.G.). H.T.K. is supported by funding from the Prostate Cancer Foundation and Swim Across America. H.I.N. receives a CNPq research fellowship. The authors acknowledge technical support from R. Karaffa and S. Durham for cell sorting.
Author information
Authors and Affiliations
Contributions
R.A., S.J.I., and J.S.H. designed and analysed the experiments. S.J.I., M.H., Jun.L., Jud.L. and T.H.N. performed experiments. S.J.I., H.T.K., M.C.B. and H.I.N. analysed microarray data. M.Y.G. performed immunofluorescence staining and M.Y.G. and R.N.G. analysed data. Q.S., H.-H.X., A.H.S., and G.J.F. contributed critical materials. R.A. and S.J.I. wrote the manuscript, with all authors contributing to writing and providing feedback.
Corresponding author
Ethics declarations
Competing interests
R.A., A.H.S. and G.J.F. hold patents and receive patent royalties related to the PD-1 inhibitory pathway. R.A., A.H.S. and G.J.F. declare no additional financial interests. The remaining authors declare no competing financial interests.
Extended data figures and tables
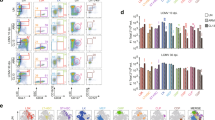
Extended Data Figure 1 LCMV GP276-specific CD8+ T cells also consist of CXCR5+ and CXCR5− CD8+ T-cell subsets during chronic infection.
a, Phenotypic analysis of GP276-specific CD8+ T cells in the spleens of immune mice that had cleared an acute LCMV Armstrong infection or mice that were chronically infected with LCMV clone 13 (day 30 after infection (p.i.)). FACS plots showing CXCR5 expression in combination with the indicated markers are gated on GP276-tetramer+ CD8+ T cells. b, Longitudinal analysis of the numbers of GP276-specific CXCR5+Tim-3− and CXCR5−Tim-3+ CD8+ T cells in the spleen at the indicated time after infection. LCMV titers in the serum are shown as the shaded yellow area. Graph shows the mean ± s.e.m. Data are the average of 8 mice from two experiments per time point (total n = 48). c, Phenotypic characterization of CXCR5+ and CXCR5− GP276-specific CD8+ T cells in the spleen of chronically infected mice (>45 days p.i.).
Extended Data Figure 2 Low-dose challenge with LCMV clone 13 results in acute infection and does not generate CXCR5+CD8+ T cells.
Mice were infected with either low-dose (2 × 102 PFU) or high-dose (2 × 106 PFU) of LCMV clone 13, and the generation of CXCR5+CD8+ T cells was examined at day 8 and day 35 p.i. a, Serum virus titers at day 8 after infection. b, Representative flow plots of CXCR5 and Tim-3 expression on GP33-specific CD8+ T cells in the spleen at day 8 and 35 p.i. Data are obtained from a total of 16 mice with 4 mice per group at each time point. Student’s t-test, where **P < 0.01.
Extended Data Figure 3 Distinct transcriptional profiles of CXCR5+ and CXCR5− CD8+ T cells from spleens of mice chronically infected with LCMV.
a, Principal component analysis of naive (CD44lo) CD8+ T cells isolated from uninfected mice and CXCR5+Tim-3−PD-1+ and CXCR5−Tim-3+PD-1+ CD8+ T cells isolated from chronically infected mice (>45 days p.i.). Each square represents an individual biological replicate. b, Relative expression of selected genes as determined by Affymetrix microarray analysis. Data are shown as fold change relative to naive (CD44lo) CD8+ T cells. Graphs show the mean ± s.e.m.
Extended Data Figure 4 LCMV-specific CXCR5+CD8+ T cells are more polyfunctional than CXCR5–CD8+ T cells.
Splenocytes from chronically infected mice (>45 days p.i.) were stimulated with GP33-41 peptide for 5 h followed by phenotypic marker staining and intracellular staining. a, Gating strategy for IFN-γ+PD-1+CXCR5+Tim-3− and IFNγ+PD-1+CXCR5−Tim-3+ CD8+ T cells. b, Representative FACS plots and graph showing the frequency of TNFα- and IL-2-producing cells among IFN-γ+CD8+ T cells. Data are representative of 2 experiments (n = 4 per experiment). Student’s paired t-test, where **P < 0.01.
Extended Data Figure 5 Analysis of reactome pathways, mTOR signalling and fatty-acid metabolism in CXCR5+ and CXCR5− CD8 T cells from LCMV chronically infected mice.
a, Reactome pathways in CXCR5+ Tim-3− PD-1+ and CXCR5− Tim-3+ PD-1+ CD8 T cells isolated from the spleens of mice chronically infected with LCMV (>45 days p.i.). GSEA (nominal P < 0.01; 1,000 permutations) was used to identify positive (red, maximum normalized enrichment score (NES) = 3.2) or negative (blue, min NES = −3.7) enrichment of Reactome pathways (http://www.reactome.org/) in CXCR5+Tim-3−PD-1+ and CXCR5− Tim-3+PD-1+ CD8 T cells using meta-analysis. The size of the circles (nodes) represents the number of genes on each pathway. The links between circles (edges) represent the number of genes shared by two given pathways. The networks were generated using Cytoscape. b, GSEA on mTOR signalling and fatty acid metabolism. Bars represent pathways with nominal P value <0.01. c, d, Splenocytes from chronically infected mice (>45 days p.i.) were stimulated with medium or GP33-41 peptide for 1 h followed by phenotypic marker staining and phosphorylated S6 ribosomal protein (pS6) staining. Flow cytometry analysis (c) and MFI (d) of pS6 expression in CXCR5+ and CXCR5− CD8+ T-cell subsets after ex vivo stimulation. Data are representative of 2 experiments (n = 4 per experiment). Student’s paired t-test, where *P < 0.05.
Extended Data Figure 6 Comparison of gene signatures of CXCR5+ and CXCR5− CD8+ T-cell subsets from chronically infected mice with Id2−/−Id3−/− innate TFH-like CD4+ T cells.
a, GSEA was performed using genes pre-ranked by the mean Z-score values of each CD8 subset (naive, CXCR5+ or CXCR5−) calculated across all samples. Splenic CD4+ TFH gene signatures from wild-type mice and thymic innate variant TFH gene signatures from Id2−/−Id3−/− mice (GSE64779) (ref. 36) were used as gene sets in our GSEA. Genes were considered up- or downregulated in cell subsets compared to control (sorted CD4+TCRβ+CD8– cells) if there was a fold-change >2 and P < 0.05 (ref. 36). b, Heat map illustrating the relative expression of the indicated genes of Id2−/−Id3−/− CD4+ T cells defined in ref. 36 compared to those of naive, CXCR5+ and CXCR5− CD8+ T cells. GSEA analysis revealed some interesting similarities and differences between the CXCR5+ CD8+ T cells from chronically infected mice and the Id2−/−Id3−/− TFH-like CD4+ T cells. These two cell populations are distinct, but share certain biological properties such as increased self-renewal activity. For example, some of the interesting inhibitory and costimulatory molecules such as Pdcd1 (PD-1), Tnfsf14 (LIGHT), Cd28, and Icos were commonly upregulated in both CXCR5+ CD8 and innate TFH-like CD4+ T cells, whereas molecules like Cd244 (2B4), Prf1, Fasl and Gzmb were downregulated in both cell types. However, there were also many differences, perhaps the most notable being the low expression of Tcf7 (TCF1) in the innate CD4+ T cells compared to the high expression of Tcf7 in the CXCR5+CD8+ T cells and the critical role of this transcription factor in the generation of these cells. Notably, the CD4+ T-cell population defined in ref. 36 is genetically deficient in both Id2 and Id3, whereas the CXCR5+CD8+ T cells express high levels of Id3. Thus, many aspects of the transcriptional program of these two cell types will be distinct.
Extended Data Figure 7 Tissue distribution of LCMV-specific CXCR5+ and CXCR5− CD8+ T cells in chronically infected mice.
a, b, Frequency (a) and numbers (b) of LCMV-specific CXCR5+ and CXCR5− CD8+ T cells in the indicated tissues at >45 days p.i. in chronically infected mice (SP, spleen; BM, bone marrow; LN, mesenteric lymph nodes; IEL, intestinal epithelial lymphocytes). c, d, Representative FACS plots (c) and graph (d) showing the frequency of CXCR5+Tim-3− GP33-specific CD8+ T cells in the blood and spleen at day 8 and day 30 p.i. FACS plots are gated on DbGP33 tetramer+ CD8+ T cells. Data are obtained from 4 or 8 mice. Graphs show the mean ± s.e.m. Student’s t-test, where **P < 0.01; *P < 0.05.
Extended Data Figure 8 Anatomic localization of CXCR5+ and CXCR5− CD8+ T cells in the spleen and in vitro migration of the two CD8+ T-cell subsets to CXCL13.
a, Analysis of TCF-1 and PD-1 expression in CD8+ T cell subsets in the spleen of chronically infected mice (>45 days p.i.). Left and middle FACS plots are gated on total CD8+ T cells and PD-1+ CD8+ T cells, respectively. Right FACS plot displays the overlay of TCF-1 and PD-1 expression in gated PD-1−, PD-1+CXCR5+Tim-3− and PD-1+CXCR5−Tim-3+ CD8+ T cells. b, c, Nuclear histocytometry analysis of the spleen (b) and frequency (c) of the CD8+ T-cell subsets within the respective zones of the spleen 10 days after LCMV clone 13 infection (n = 3). d, Representative immunofluorescence staining of LCMV clone 13 infection in the spleen (>45 days p.i.). The spleen was stained for LCMV antigen (red), IgD (blue) and CD3 (white) and examined via microscopy (n = 4). Note, LCMV infection is mostly in the red pulp. e, CXCR5 expression on splenic naive (CD44lo) CD8+ T cells, B cells and CXCR5+ GP33-specific CD8+ T cells from chronically infected mice (>45 days p.i.). f, Relative migration of sorted B cells and CXCR5+Tim-3−PD-1+ and CXCR5−Tim-3+PD-1+ CD8+ T-cell subsets in response to CXCL13, which is a ligand for CXCR5. Data are combined from two experiments performed in duplicate wells for each sample. Graphs show the mean and s.e.m. Student’s t-test, where **P < 0.01; *P < 0.05.
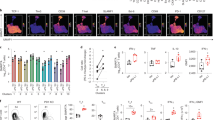
Extended Data Figure 9 CXCR5+CD8+ T cells selectively undergo proliferation after LCMV clone 13 challenge.
a, Sorted PD-1+CXCR5+Tim-3− and PD-1+CXCR5−Tim-3+ CD8+ T cells isolated from CD45.2+ chronically infected mice (>45 days p.i.) were adoptively transferred into naive CD45.1+ recipient mice, followed by LCMV-clone- 13 challenge. Two sets of adoptive-transfer experiments were performed; one using a low dose of donor cells (2,500 cells) and another with a large dose of donor cells (90,000 cells). The data shown in Fig. 3d, e are from the high-dose transfer experiment. Data in b–d are from the high-dose transfer; and in e–h are from the low-dose transfer. b, Expansion of CXCR5+CD8+ T cells in the blood after LCMV clone 13 infection. c, Number of cells in the spleen and liver 14 days after infection. d, Phenotypic analysis of transferred donor CXCR5+Tim-3− cells in the spleen 14 days after challenge, showing proliferation and differentiation of this subset. Data are representative of 3 experiments (n = 4 or 6 mice per experiment). e, Expansion of CXCR5+CD8+ T cells (low-dose transfer) in the blood after infection with LCMV clone 13. f, Number of cells in the spleen and liver 14 days after challenge. g, Phenotypic analysis of transferred donor CXCR5+Tim-3− and CXCR5−Tim-3+ cells in the spleen and liver at 14 days after infection showing differentiation of CXCR5+CD8+ T cells. h, Number of CXCR5+Tim-3− and CXCR5−Tim-3+ CD8+ T cells derived from donor CXCR5+ or CXCR5− CD8+ T cells in the spleen and liver after clone 13 challenge. Dashed line indicates the limit of detection. Data are combined from two experiments (n = 8 or 10 per group, total n = 18). Graph shows the mean and s.e.m. Student’s t-test, where **P < 0.01.
Extended Data Figure 10 Enhanced conversion of CXCR5+Tim-3−CD8+ T cells to Tim-3+CD8+ T cells after PD-1 blockade.
a, Sorted PD-1+CXCR5+Tim-3− and PD-1+CXCR5−Tim-3+ CD8+ T cells isolated from CD45.2+ chronically infected mice (>45 days p.i.) were adoptively transferred into infection-matched CD45.1+ recipient mice, followed by treatment with anti-PD-L1 antibody. b, Phenotypic analysis of sorted donor CD8+ T-cell subsets before transfer and 14 days after the transfer followed by PD-1 blockade. Data are representative of 2 experiments (total n = 5, 7, or 9 mice per group).
Rights and permissions
About this article
Cite this article
Im, S., Hashimoto, M., Gerner, M. et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). https://doi.org/10.1038/nature19330
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19330
This article is cited by
-
Association of lymphocyte subsets with the efficacy and prognosis of PD‑1 inhibitor therapy in advanced gastric cancer: results from a monocentric retrospective study
BMC Gastroenterology (2024)
-
Per-cell histone acetylation is associated with terminal differentiation in human T cells
Clinical Epigenetics (2024)
-
Id2 epigenetically controls CD8+ T-cell exhaustion by disrupting the assembly of the Tcf3-LSD1 complex
Cellular & Molecular Immunology (2024)
-
PD-1 defines a distinct, functional, tissue-adapted state in Vδ1+ T cells with implications for cancer immunotherapy
Nature Cancer (2024)
-
FOXP1 and KLF2 reciprocally regulate checkpoints of stem-like to effector transition in CAR T cells
Nature Immunology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.