Abstract
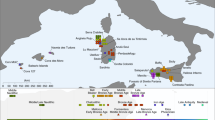
We report genome-wide ancient DNA from 44 ancient Near Easterners ranging in time between ~12,000 and 1,400 bc, from Natufian hunter–gatherers to Bronze Age farmers. We show that the earliest populations of the Near East derived around half their ancestry from a ‘Basal Eurasian’ lineage that had little if any Neanderthal admixture and that separated from other non-African lineages before their separation from each other. The first farmers of the southern Levant (Israel and Jordan) and Zagros Mountains (Iran) were strongly genetically differentiated, and each descended from local hunter–gatherers. By the time of the Bronze Age, these two populations and Anatolian-related farmers had mixed with each other and with the hunter–gatherers of Europe to greatly reduce genetic differentiation. The impact of the Near Eastern farmers extended beyond the Near East: farmers related to those of Anatolia spread westward into Europe; farmers related to those of the Levant spread southward into East Africa; farmers related to those of Iran spread northward into the Eurasian steppe; and people related to both the early farmers of Iran and to the pastoralists of the Eurasian steppe spread eastward into South Asia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
European Nucleotide Archive
Data deposits
The aligned sequences are available through the European Nucleotide Archive under accession number PRJEB14455. Fully public subsets of the analysis datasets are at http://genetics.med.harvard.edu/reichlab/Reich_Lab/Datasets.html. The complete dataset (including present-day humans for which the informed consent is not consistent with public posting of data) is available to researchers who send a signed letter to D.R. indicating that they will abide by specified usage conditions (Supplementary Information, section 2).
References
Barker, G. & Goucher, C. The Cambridge World History Volume II: A World with agriculture, 12,000 BCE–500 CE (Cambridge Univ. Press, 2015)
Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. The History and Geography of Human Genes. (Princeton Univ. Press, 1994)
Gamba, C. et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat. Commun. 5, 5257 (2014)
Pinhasi, R. et al. Optimal ancient DNA yields from the inner ear part of the human petrous bone. PLoS One 10, e0129102 (2015)
Fu, Q. et al. DNA analysis of an early modern human from Tianyuan Cave, China. Proc. Natl Acad. Sci. USA 110, 2223–2227 (2013)
Fu, Q. et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015)
Haak, W. et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015)
Mathieson, I. et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015)
Jones, E. R. et al. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nat. Commun. 6, 8912 (2015)
Allentoft, M. E. et al. Population genomics of Bronze Age Eurasia. Nature 522, 167–172 (2015)
Fu, Q. et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449 (2014)
Günther, T. et al. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc. Nat. Acad Sci. USA (2015)
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014)
Olalde, I. et al. A common genetic origin for early farmers from Mediterranean Cardial and Central European LBK cultures. Mol. Biol. Evol. 32, 3132–3142 (2015)
Raghavan, M. et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505, 87–91 (2014)
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012)
Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006)
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009)
Prufer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014)
Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012)
Wall, J. D. et al. Higher levels of neanderthal ancestry in East Asians than in Europeans. Genetics 194, 199–209 (2013)
Green, R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010)
Brace, C. L. et al. The questionable contribution of the Neolithic and the Bronze Age to European craniofacial form. Proc. Natl Acad. Sci. USA 103, 242–247 (2006)
Ferembach, D. Squelettes du Natoufien d’Israel., etude anthropologique. Anthropologie 65, 46–66 (1961)
Fadhlaoui-Zid, K. et al. Genome-wide and paternal diversity reveal a recent origin of human populations in North Africa. PLoS One 8, e80293 (2013)
Henn, B. M. et al. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 8, e1002397 (2012)
Bhatia, G., Patterson, N., Sankararaman, S. & Price, A. L. Estimating and interpreting FST: the impact of rare variants. Genome Res. 23, 1514–1521 (2013)
Fernández, E. et al. Ancient DNA analysis of 8000 B.C. near eastern farmers supports an early neolithic pioneer maritime colonization of Mainland Europe through Cyprus and the Aegean Islands. PLoS Genet. 10, e1004401 (2014)
Ammerman, A. J., Pinhasi, R. & Banffy, E. Comment on Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science 312, 1875; author reply 1875 (2006)
Pagani, L. et al. Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am. J. Hum. Genet. 91, 83–96 (2012)
Pickrell, J. K. et al. Ancient west Eurasian ancestry in southern and eastern Africa. Proc. Natl Acad. Sci. USA 111, 2632–2637 (2014)
Keller, A. et al. New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat Commun 3, 698 (2012)
Moorjani, P. et al. Genetic evidence for recent population mixture in India. Am. J. Hum. Genet. 93, 422–438 (2013)
Reich, D., Thangaraj, K., Patterson, N., Price, A. L. & Singh, L. Reconstructing Indian population history. Nature 461, 489–494 (2009)
Fu, Q. et al. The genetic history of Ice Age Europe. Nature 534, 200–205 (2016)
Dabney, J. et al. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl Acad. Sci. USA 110, 15758–15763 (2013)
Rohland, N., Harney, E., Mallick, S., Nordenfelt, S. & Reich, D. Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Phil. Trans. R. Soc. Lond. B 370, 20130624 (2015)
Briggs, A. W. et al. Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 38, e87 (2010)
Korlević, P. et al. Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. Biotechniques 59, 87–93 (2015)
Meyer, M. et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 505, 403–406 (2014)
Behar, D. M. et al. A “Copernican” reassessment of the human mitochondrial DNA tree from its root. Am. J. Hum. Genet. 90, 675–684 (2012)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Korneliussen, T. S., Albrechtsen, A. & Nielsen, R. ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014)
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015)
Busing, F. T. A., Meijer, E. & Leeden, R. Delete-m Jackknife for Unequal m. Stat. Comput. 9, 3–8 (1999)
Sudmant, P. H. et al. Global diversity, population stratification, and selection of human copy-number variation. Science 349, aab3761 (2015)
Reich, D. et al. Reconstructing Native American population history. Nature 488, 370–374 (2012)
Gallego Llorente, M. et al. Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa. Science 350, 820–822 (2015)
Acknowledgements
We thank the 238 human subjects who donated samples for genome-wide analysis, and D. Labuda and P. Zalloua for sharing samples from Poland and Lebanon. The Fig. 1a map was plotted in R using the worldHiRes map of the ‘mapdata’ package (using public domain data from the CIA World Data Bank II). We thank O. Bar-Yosef, M. Bonogofsky, I. Hershkowitz, M. Lipson, I. Mathieson, H. May, R. Meadow, I. Olalde, S. Paabo, P. Skoglund, and N. Nakatsuka for comments and critiques, and D. Bradley, M. Dallakyan, S. Esoyan, M. Ferry and M. Michel, and A. Yesayan, for contributions to bone preparation and ancient DNA work. D.F. and M.N. were supported by Irish Research Council grants GOIPG/2013/36 and GOIPD/2013/1, respectively. S.C. was funded by the Irish Research Council for Humanities and Social Sciences (IRCHSS) ERC Support Programme. Q.F. was funded by the Bureau of International Cooperation of the Chinese Academy of Sciences, the National Natural Science Foundation of China (L1524016) and the Chinese Academy of Sciences Discipline Development Strategy Project (2015-DX-C-03). The Scottish diversity data was funded by the Chief Scientist Office of the Scottish Government Health Directorates (CZD/16/6), the Scottish Funding Council (HR03006), and a project grant from the Scottish Executive Health Department, Chief Scientist Office (CZB/4/285). M.S., A.Tön., M.B. and P.K. were supported by the German Research Foundation (CRC 1052; B01, B03, C01). M.S.-P. was funded by a Wenner-Gren Foundation Dissertation Fieldwork grant (9005), and by the National Science Foundation DDRIG (BCS-1455744). P.K. was funded by the Federal Ministry of Education and Research, Germany (FKZ: 01EO1501). J.F.W. acknowledge the MRC ‘QTL in Health and Disease’ programme grant. The Romanian diversity data was supported by the EC Commission, Directorate General XII (Supplementary Agreement ERBCIPDCT 940038 to the Contract ERBCHRXCT 920032, coordinated by A. Piazza, Turin, Italy). M.R. received support from the Leverhulme Trust’s Doctoral Scholarship programme. O.S. and A.Tor. were supported by the University of Pavia (MIGRAT-IN-G) and the Italian Ministry of Education, University and Research: Progetti Ricerca Interesse Nazionale 2012. The Raqefet Cave Natufian project was supported by funds from the National Geographic Society (grant 8915-11), the Wenner-Gren Foundation (grant 7481) and the Irene Levi-Sala CARE Foundation, while radiocarbon dating on the samples was funded by the Israel Science Foundation (grant 475/10; E. Boaretto). R.P. was supported by ERC starting grant ADNABIOARC (263441). D.R. was supported by NIH grant GM100233, by NSF HOMINID BCS-1032255, and is a Howard Hughes Medical Institute investigator.
Author information
Authors and Affiliations
Contributions
R.P. and D.R. conceived the idea for the study. D.N., G.R., D.C.M., S.C., S.A.R., G.L., F.B., B.Gas., J.M.M., M.G., V.E., A.M., C.M., F.G., N.A.H. and R.P. assembled skeletal material. N.R., D.F., M.N., B.Gam., K.Si., S.C., K.St., E.H., Q.F., G.G.-F., E.R.J., R.P. and D.R. performed or supervised ancient DNA wet laboratory work. L.B, M.B., A.C., G.C., D.C., P.F., E.G., S.M.K., P.K., J.K., D.M., M.M., D.A.M., S.O., M.B.R., O.S., M.S.-P., G.S., M.S., A.Tön., A.Tor., J.F.W., L.Y. and D.R. assembled present-day samples for genotyping. I.L, N.P. and D.R. developed methods for data analysis. I.L., S.M., Q.F., N.P. and D.R. analysed data. I.L., R.P. and D.R. wrote the manuscript and supplements.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks O. Bar-Yosef, G. Coop and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 2 Genetic structure in ancient West Eurasian populations across time and decline of genetic differentiation over time.
a, ADMIXTURE model-based clustering analysis of 2,583 present-day humans and 281 ancient samples; we show the results only for ancient samples for K = 11 clusters. b, Pairwise FST between 19 Ancient West Eurasian populations (arranged in approximate chronological order), and select present-day populations.
Extended Data Figure 3 Outgroup f3(Mbuti; X, Y) for pairs of ancient populations.
The dendrogram is plotted for convenience and should not be interpreted as a phylogenetic tree. Areas of high shared genetic drift are ‘yellow’ and include from top-right to bottom-left along the diagonal: early Anatolian and European farmers; European hunter–gatherers, Steppe populations and populations admixed with steppe ancestry; populations from the Levant from the Epipalaeolithic (Natufians) to the Bronze Age; populations from Iran from the Mesolithic to the Late Neolithic.
Extended Data Figure 4 Reduction of genetic differentiation in West Eurasia over time.
We measure differentiation by FST. Each column of the 5 × 5 matrix of plots represents a major region and each row the earliest population with at least two individuals from each major region.
Extended Data Figure 5 West Eurasian related admixture in East Africa, Eastern Eurasia and South Asia.
a, Levantine ancestry in Eastern Africa in the Human Origins dataset. b, Levantine ancestry in different Eastern African population in the dataset from Pagani et al. (2012); the remainder of the ancestry is a clade with Mota, a ~4,500 year old sample from Ethiopia49. c, EHG ancestry in Eastern Eurasians. d, Afontova Gora (AG2)-related ancestry in Eastern Eurasians; the remainder of their ancestry is a clade with Onge. e, Mixture proportions for South Asian populations showing that they can be modelled as having West Eurasian-related ancestry similar to that in populations from both the Eurasian steppe and Iran.
Extended Data Figure 6 Inferred position of ancient populations in West Eurasian PCA according to the model of Fig. 4.
Extended Data Figure 7 Admixture from ghost populations using ‘cline intersection’.
a–f, We model each Test population (purple) as a mixture (pink) of a fixed reference population (blue) and a ghost population (orange) residing on the cline defined by two other populations (red and green) according to the visualization method of Supplementary Information, section 10. a, Early/Middle Bronze Age steppe populations are a mixture of Iran_ChL and a population on the WHG→SHG cline. b, Scandinavian hunter–gatherers (SHG) are a mixture of WHG and a population on the Iran_ChL→Steppe_EMBA cline. c, Caucasus hunter–gatherers (CHG) are a mixture of Iran_N and both WHG and EHG. d, Late Neolithic/Bronze Age Europeans are a mixture of the preceding Europe_MNChL population and a population with both EHG and Iran_ChL ancestry. e, Somali are a mixture of Mota49 and a population on the Iran_ChL→Levant_BA cline. f, Eastern European hunter–gatherers (EHG) are a mixture of WHG and a population on the Onge→Han cline.
Extended Data Figure 8 Admixture from a ‘ghost’ ANE population into both European and Eastern Eurasian ancestry.
EHG, and Upper Palaeolithic Siberians Mal’ta 1 (MA1) and Afontova Gora 2 (AG2) are positioned near the intersection of clines formed by European hunter–gatherers (WHG, SHG, EHG) and Eastern non-Africans in the space of outgroup f3-statistics of the form f3(Mbuti; Papuan, Test) and f3(Mbuti; Switzerland_HG, Test).
Supplementary information
Supplementary Table 1
This file contains Supplementary Data Table 1. (XLSX 65 kb)
Supplementary Table 2
This file contains Supplementary Data Table 2. (XLSX 23 kb)
Supplementary Table 3
This file contains Supplementary Data Table 3. (XLSX 60 kb)
Supplementary Information
This file contains Supplementary Text, Data and References – see contents page for details. (PDF 10045 kb)
Rights and permissions
About this article
Cite this article
Lazaridis, I., Nadel, D., Rollefson, G. et al. Genomic insights into the origin of farming in the ancient Near East. Nature 536, 419–424 (2016). https://doi.org/10.1038/nature19310
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature19310
This article is cited by
-
Evolutionary origin of germline pathogenic variants in human DNA mismatch repair genes
Human Genomics (2024)
-
Kinship practices at the early bronze age site of Leubingen in Central Germany
Scientific Reports (2024)
-
Koban culture genome-wide and archeological data open the bridge between Bronze and Iron Ages in the North Caucasus
European Journal of Human Genetics (2024)
-
The Allen Ancient DNA Resource (AADR) a curated compendium of ancient human genomes
Scientific Data (2024)
-
Population genomics of post-glacial western Eurasia
Nature (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.