Abstract
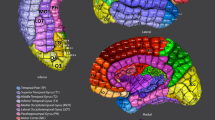
Understanding the amazingly complex human cerebral cortex requires a map (or parcellation) of its major subdivisions, known as cortical areas. Making an accurate areal map has been a century-old objective in neuroscience. Using multi-modal magnetic resonance images from the Human Connectome Project (HCP) and an objective semi-automated neuroanatomical approach, we delineated 180 areas per hemisphere bounded by sharp changes in cortical architecture, function, connectivity, and/or topography in a precisely aligned group average of 210 healthy young adults. We characterized 97 new areas and 83 areas previously reported using post-mortem microscopy or other specialized study-specific approaches. To enable automated delineation and identification of these areas in new HCP subjects and in future studies, we trained a machine-learning classifier to recognize the multi-modal ‘fingerprint’ of each cortical area. This classifier detected the presence of 96.6% of the cortical areas in new subjects, replicated the group parcellation, and could correctly locate areas in individuals with atypical parcellations. The freely available parcellation and classifier will enable substantially improved neuroanatomical precision for studies of the structural and functional organization of human cerebral cortex and its variation across individuals and in development, aging, and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Brodmann, K. Vergleichende Lokalisationslehre der Grosshirnrinde in ihren Prinzipien dargestellt auf Grund des Zellenbaues (J. A. Barth, 1909); Brodmann’s Localization in the Cerebral Cortex (Smith Gordon, 1994) [transl. Garey, L.J.]
Felleman, D. J. & Van Essen, D. C. Distributed hierarchical processing in the primate cerebral cortex. Cereb. Cortex 1, 1–47 (1991)
Nieuwenhuys, R. The myeloarchitectonic studies on the human cerebral cortex of the Vogt–Vogt school, and their significance for the interpretation of functional neuroimaging data. Brain Struct. Funct. 218, 303–352 (2013)
Van Essen, D. C., Glasser, M. F., Dierker, D. L., Harwell, J. & Coalson, T. Parcellations and hemispheric asymmetries of human cerebral cortex analyzed on surface-based atlases. Cereb. Cortex 22, 2241–2262 (2012)
Glasser, M. F. et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124 (2013)
Smith, S. M. et al. Resting-state fMRI in the Human Connectome Project. Neuroimage 80, 144–168 (2013)
Uğurbil, K. et al. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage 80, 80–104 (2013)
Van Essen, D. C. et al. The WU-Minn Human Connectome Project: an overview. Neuroimage 80, 62–79 (2013)
Glasser, M. F., Goyal, M. S., Preuss, T. M., Raichle, M. E. & Van Essen, D. C. Trends and properties of human cerebral cortex: correlations with cortical myelin content. Neuroimage 93, 165–175 (2014)
Glasser, M. F. & Van Essen, D. C. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J. Neurosci. 31, 11597–11616 (2011)
Barch, D. M. et al. Function in the human connectome: task-fMRI and individual differences in behavior. Neuroimage 80, 169–189 (2013)
Caspers, S., Eickhoff, S. B., Zilles, K. & Amunts, K. Microstructural grey matter parcellation and its relevance for connectome analyses. Neuroimage 80, 18–26 (2013)
Schleicher, A., Amunts, K., Geyer, S., Morosan, P. & Zilles, K. Observer-independent method for microstructural parcellation of cerebral cortex: a quantitative approach to cytoarchitectonics. Neuroimage 9, 165–177 (1999)
Robinson, E. C. et al. MSM: a new flexible framework for multimodal surface matching. Neuroimage 100, 414–426 (2014)
Zilles, K. & Amunts, K. Centenary of Brodmann’s map—conception and fate. Nat. Rev. Neurosci. 11, 139–145 (2010)
Cohen, A. L. et al. Defining functional areas in individual human brains using resting functional connectivity MRI. Neuroimage 41, 45–57 (2008)
Kolster, H., Peeters, R. & Orban, G. A. The retinotopic organization of the human middle temporal area MT/V5 and its cortical neighbors. J. Neurosci. 30, 9801–9820 (2010)
Wang, L., Mruczek, R. E., Arcaro, M. J. & Kastner, S. Probabilistic maps of visual topography in human cortex. Cereb. Cortex 25, 3911–3931 (2015)
Gordon, E. M. et al. Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex 26, 288–303 (2016)
Shen, X., Tokoglu, F., Papademetris, X. & Constable, R. T. Groupwise whole-brain parcellation from resting-state fMRI data for network node identification. Neuroimage 82, 403–415 (2013)
Yeo, B. T. et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011)
Hopf, A. Uber die Verteilung myeloarchitektonischer Merkmale in der Stirnhirnrinde beim Menschen. J. Hirnforsch. 2, 311–333 (1956)
Van Essen, D. C. & Glasser, M. F. In vivo architectonics: a cortico-centric perspective. Neuroimage 93, 157–164 (2014)
Olman, C. A. et al. Layer-specific fMRI reflects different neuronal computations at different depths in human V1. PLoS One 7, e32536 (2012)
Polimeni, J. R., Fischl, B., Greve, D. N. & Wald, L. L. Laminar analysis of 7T BOLD using an imposed spatial activation pattern in human V1. Neuroimage 52, 1334–1346 (2010)
Yacoub, E., Harel, N. & Ugurbil, K. High-field fMRI unveils orientation columns in humans. Proc. Natl Acad. Sci. USA 105, 10607–10612 (2008)
Zimmermann, J. et al. Mapping the organization of axis of motion selective features in human area MT using high-field fMRI. PLoS One 6, e28716 (2011)
Smith, S. M. et al. Functional connectomics from resting-state fMRI. Trends Cogn. Sci. 17, 666–682 (2013
Hacker, C. D. et al. Resting state network estimation in individual subjects. Neuroimage 82, 616–633 (2013)
Tavor, I. et al. Task-free MRI predicts individual differences in brain activity during task performance. Science 352, 216–220 (2016)
Van Essen, D. C. et al. The brain analysis library of spatial maps and atlases (BALSA) database. Neuroimage http://dx.doi.org/10.1016/j.neuroimage.2016.04.002 (2016)
Hill, J. et al. A surface-based analysis of hemispheric asymmetries and folding of cerebral cortex in term-born human infants. J. Neurosci. 30, 2268–2276 (2010)
Van Essen, D. C. & Dierker, D. L. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron 56, 209–225 (2007)
Glasser, M. F. et al. The Human Connectome Project’s neuroimaging approach. Nat. Neuroscience (in press)
Fischl, B. et al. Automatically parcellating the human cerebral cortex. Cereb. Cortex 14, 11–22 (2004)
Fischl, B. FreeSurfer. NeuroImage 62, 774–781 (2012)
Filippini, N. et al. Distinct patterns of brain activity in young carriers of the APOE-ε4 allele. Proc. Natl Acad. Sci. USA 106, 7209–7214 (2009)
Abdollahi, R. O. et al. Correspondences between retinotopic areas and myelin maps in human visual cortex. Neuroimage 99, 509–524 (2014)
Griffanti, L. et al. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. Neuroimage 95, 232–247 (2014)
Salimi-Khorshidi, G. et al. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage 90, 449–468 (2014)
Caspers, S. et al. The human inferior parietal lobule in stereotaxic space. Brain Struct. Funct. 212, 481–495 (2008)
Malikovic, A. et al. Cytoarchitectonic analysis of the human extrastriate cortex in the region of V5/MT+: a probabilistic, stereotaxic map of area hOc5. Cereb. Cortex 17, 562–574 (2007)
Acknowledgements
We thank the members of the WU-Minn-Ox HCP Consortium for invaluable contributions to data acquisition, analysis, and sharing and E. Reid and S. Danker for assistance with preparing the manuscript. Supported by NIH F30 MH097312 (M.F.G.), ROIMH-60974 (D.C.V.E.), NIH F30 MH099877 (C.D.H.), the Human Connectome Project grant (1U54MH091657) from the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research, and the Wellcome Trust Strategic Award 098369/Z/12/Z (S.M.S., J.A., C.F.B., M.J.).
Author information
Authors and Affiliations
Contributions
M.F.G. and D.C.V.E. designed the study and carried out the analyses. M.F.G., T.S.C., E.C.R., C.D.H., J.H., E.Y., K.U., J.A., C.F.B., M.J., and S.M.S. contributed novel methods. M.F.G., T.S.C., E.C.R., C.D.H., E.Y., J.A., C.F.B., M.J., S.M.S., and D.C.V.E. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks R. Poldrack, F. Tong and T. Yeo for their contribution to the peer review of this work.
Supplementary information
Supplementary Methods
This file explains the experimental methods in detail. This information will be of particular interest for methods oriented neuroimaging scientists interested in exactly what was done and why. It contains 11 Supplementary figures. (PDF 4565 kb)
Supplementary Results and Discussion
This file contains 12 supplementary figures and supplementary text expanding on the reproducibility of the data used to generate the parcellation, cross validation of parcellation, an exploration of atypical parcellations in individual subjects, a peek inside the areal classifier, and a discussion. (PDF 6176 kb)
Supplementary Neuroanatomical Results
This file provides a detailed neuroanatomical description of how each border between each pair of cortical areas was delineated and how each area was identified and named. This information will be of particular interest to neuroanatomists. It contains 25 figures and 3 tables. (PDF 13162 kb)
Rights and permissions
About this article
Cite this article
Glasser, M., Coalson, T., Robinson, E. et al. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016). https://doi.org/10.1038/nature18933
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18933
This article is cited by
-
Improved cognition after rifaximin treatment is associated with changes in intra- and inter-brain network functional connectivity
Journal of Translational Medicine (2024)
-
Integrative omics analysis reveals epigenomic and transcriptomic signatures underlying brain structural deficits in major depressive disorder
Translational Psychiatry (2024)
-
Spectrotemporal cues and attention jointly modulate fMRI network topology for sentence and melody perception
Scientific Reports (2024)
-
Driving and suppressing the human language network using large language models
Nature Human Behaviour (2024)
-
Nuclei-specific hypothalamus networks predict a dimensional marker of stress in humans
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.