Abstract
Using brain atlases to localize regions of interest is a requirement for making neuroscientifically valid statistical inferences. These atlases, represented in volumetric or surface coordinate spaces, can describe brain topology from a variety of perspectives. Although many human brain atlases have circulated the field over the past fifty years, limited effort has been devoted to their standardization. Standardization can facilitate consistency and transparency with respect to orientation, resolution, labeling scheme, file storage format, and coordinate space designation. Our group has worked to consolidate an extensive selection of popular human brain atlases into a single, curated, open-source library, where they are stored following a standardized protocol with accompanying metadata, which can serve as the basis for future atlases. The repository containing the atlases, the specification, as well as relevant transformation functions is available in the neuroparc OSF registered repository or https://github.com/neurodata/neuroparc.
Similar content being viewed by others
Introduction
Understanding the brain’s organization is one of the key challenges in human neuroscience1 and is critical for clinical translation2. Parcellation of the brain into functionally and structurally distinct regions has seen impressive advances in recent years3, and has grown the field of network neuroscience4,5. Through a range of techniques such as clustering6,7,8,9, multivariate decomposition10,11, gradient based connectivity1,12,13,14,15,16, and multimodal neuroimaging1, parcellations have enabled fundamental insights into the brain’s topological organization and network properties17. In turn, these properties have allowed researchers to investigate brain-behavioral associations with developmental18,19, cognitive20,21, and clinical phenotypes22,23,24.
More recently, researchers interested in understanding brain organization are presented with a variety of brain atlases that can be used to define nodes of network-based analyses25. While this variety is a boon to researchers, the use of different parcellations across studies makes assessing reproducibility of brain-behavior relationships difficult (e.g. comparing across parcellations with different organizations and numbers of nodes5). Amalgamating multiple brain parcellations into a single, standardized, curated list would offer researchers a valuable resource for evaluating replication of neuroimaging studies.
Some efforts to consolidate these atlases is already underway. For example, Nilearn is a popular Python package that provides machine-learning and informatics tools for neuroimaging26. Nilearn provides several single line command line interface functions to ‘fetch’ both atlases and datasets. Nilearn includes twelve anatomically and functionally defined atlases, such as the Harvard-Oxford27 and Automated Anatomical Labeling (AAL)28 parcellation. Although a promising prototype, Nilearn’s current atlas collection represent a limited range of available atlases, and the more recent gradient based, surface based, and multimodal parcellations have yet to be included into any central repository. More importantly, existing atlas repositories have not attempted to systematically standardize their collections following a single specification. Without well-established standards, the investigator is faced with limited information about each atlas, so connecting neuroscientific findings to the organization of the atlas becomes more difficult. Moreover, if the investigator requires a comparison across atlases, some form of metadata must be available that describes the similarities and differences between them. Neuroparc mitigates these issues by providing: (1) a detailed atlas specification which will enable researchers to both easily understand existing atlases and generate new atlases compliant with this specification, (2) a repository of the most commonly used atlases in neuroimaging, all stored in that specification, and (3) a set of functions for transforming, comparing, and visualizing these atlases. The Neuroparc package presented here includes 46 different adult human brain parcellations—including surface-based and volume-based. Here, we provide an overview of the relationship between these parcellations via comparison of the spatial similarity between atlases, as measured by Dice coefficient and adjusted mutual information. To facilitate replication and extension of this work, all the data and code are available from a the registered OSF repository29 or the github repository https://github.com/neurodata/neuroparc.
Results
Atlases
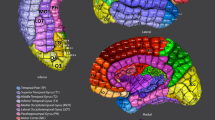
Through the use of the python scripts provided in Neuroparc, 46 atlases were resampled to either 1 mm3, 2 mm3, or 4 mm3 and registered to the Montreal Neurological Institute 152 Nonlinear 6th generation reference brain (MNI). Each atlas had a accompanying JSON file containing the relevant metadata described in Methods. Of the 46 atlases, 17 lost at least one ROI from the resampling and registration process, which is recorded in the JSON files. This phenomenon was more common with atlases containing a greater amount of ROIs due to the smaller average ROI size. The smaller the ROI, the greater the chance it is overwritten by surrounding larger ROIs when down-sampled or registered. Visualizations of a variety of atlases at 1 mm³ resolution are shown in Fig. 1. The number of ROIs present at each voxel size is recorded in Table 1
A comparison of the regions present in the major atlases available in Neuroparc. These visualizations were made using MIPAV tri-planar views on the same slice numbers. Each atlas shows a cross-section in each of the canonical orthogonal planes (H = Horizontal, S = Sagittal, C = Coronal). For most atlases, the slice numbers were (90, 108, 90). There are a few exceptions for visualization purposes: JHU: (90, 108, 109), Slab907: (95, 104, 95), Slab1068: (93, 105, 93)1,27,28,32,35,36,38,39,52,53,54.
Dice coefficient
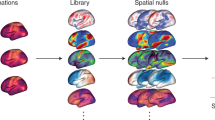
A python script, provided in Neuroparc using functions from both AFNI30 and FSL31, was used to calculate the Dice Coefficients between pairs of atlases. See the Dice Coefficient section in Methods for more information about the calculation. Dice Score Maps, such as Fig. 2, for each of the pairs of atlases can be found on the Neuroparc OSF registered repository29. The purpose of these maps are to both reinforce the differences between each parcellation and what they represent, as well as serve as comparative metric for ROIs across atlases. Access to these values allow for easy cross-parcellation analysis, a useful tool to anyone using Neuroparc. On inspection, each Dice Coefficient Map was accurate in its representation of overlap present between ROIs of two different atlases.
Dice Score Map between the Yeo-17 Networks atlas and the 300 parcellation Schaefer atlas. In a Dice Score Map, the larger the Dice score, the larger the percentage of overlap. Due to Schaefer’s larger quantity of ROIs, several different ROIs overlap a single Yeo-17 ROI. In this Dice Map, the 0 ROI for Yeo-17 represents the background of the image, or the empty space in the image. This ROI not having a Dice value of 0 indicates that both atlases don’t cover the same amount of brain volume.
Adjusted mutual information
Using another python script, provided in Neuroparc GitHub repository, the Adjusted Mutual Information (AMI) between atlases was calculated. See the Adjusted Mutual Information section in Methods for more information about the calculation. The results, displayed in Fig. 3, affirm the necessity for having access to multiple different parcellation methods during data analysis. Each atlas was created using different reference data and designed to track specific structure or functionality present in the brain. If atlases were similar to the point of being interchangeable, this wide variety of AMI scores would not exist, along with the reason for having a repository of atlases. The AMI amongst the Schaefer atlas set32, DS atlas set33, and Slab34 atlases was consistently greater than 0.8, an expected result due to their creation using the same methodologies with different parameters and tolerances.
Both the JHU35 and Princeton36 atlases displayed a consistently low (<0.3) AMI value for the majority of other atlases. This can be explained by the fact that both atlases only relate to anatomical sub-structures, such as the visual cortex for JHU and hippocampal region for Princeton. Their limited coverage of the brain results in less mutual information with the other surface-based or volume-based atlases.
Discussion
Why use neuroparc
The purpose of the Neuroparc atlas collection and metadata formatting method is two fold: (1) to provide a repository of standardized parcellations that can be used interchangeably without any additional effort, and (2) to document all relevant information about each parcellation for easy use in research. Neuroparc succeeds in both of these aspects, as well as enabling a new level of comparison between atlases. Using the formatting method proposed in this paper, any user of the repository has the ability to find where each atlas came from, how many different ROIs exist in the atlas, the location and size of each of each ROI, how the segmentation of an atlas compares to others, and whether there is a significant correlation between areas covered by ROIs from different atlases. The formatting method also allows for constant improvement and refinement of atlas metadata, discussed below in Future Development. By standardizing the atlases, researchers can easily analyze MRI data in MNI space using a variety of atlases without additional processing. Metrics provided by Neuroparc, such as adjusted mutual information and the dice coefficient, also inform users as to how the atlases are related.
Potential issues
The method used to generate the atlases for Neuroparc can result in the loss of ROIs due to down-sampling and registration. The chance of this occurring inversely correlates with the average size of ROIs in a given atlas. The ROIs that are lost are still cataloged in the corresponding JSON file, with a value of “null” being given for the center coordinates and size. While there do exist ways to attempt to prevent this loss of information, excessive manipulation of a given atlas depending on the voxel size potentially compromises any conclusions derived using said atlas. As such, the parcellations in Neuroparc do not incorporate these additional methods and it us on the user to decide how best to resample the corresponding 1 mm voxel parcellation to fit their unique needs.
Future development
With the current iteration of Neuroparc there are several routes for improvement. The most apparent is the expansion of the atlas collection. Our proposed methodology for standardizing new atlases and tracking metadata makes this task a simple one. With the emphasis on clear and concise information, approval of any new set of atlases is a quick and simple process. There also exists the ability to standardize all atlases to other spaces besides MNI, allowing for atlases offered in both different voxel sizes and standardized spaces. Another route for growth of Neuroparc is the anatomical labeling of atlases whose ROIs do not have clearly defined anatomical boundaries. The anatomical labels currently in Neuroparc are taken from the published work where they were first made. To keep the rational of the original authors, very little was done to the labels provided, mainly rewording for clarity and to follow the largest structure to smallest structure method. Due to subjective nature of labeling the atlases, an agreed upon anatomical labeling reference would first have to be made. From there, anatomical labeling could be assigned pragmatically.
The methodology proposed in this paper, as well as the atlas repository in Neuroparc, attempt to address the lack of standardization and centralization for brain atlases. We believe that Neuroparc exemplifies the first steps towards solving this issue. We call on other researchers to utilize the resources contained within and encourage everyone to contribute.
Methods
Data compilation
The atlases contained in Neuroparc were collected from a variety of locations. As previously noted, there is no current standard for atlas storage, so all gathered datasets are converted into a single format. Collected atlases were re-sampled using AFNI’s 3dresample30 to either 1 mm3, 2 mm3, or 4 mm3 voxel resolution and then registered to a reference T1-weighted image described below. The sources and additional information for each of the atlases can be found in the README file in the GitHub repository.
Reference brain
To allow direct comparison between different atlases, a standard reference brain must be used for all involved atlases. Within Neuroparc, a single reference brain is provided and multiple resolutions, yielding a consistent coordinate space. Neuroparc uses Montreal Neurological Institute 152 Nonlinear 6th generation reference brain, abbreviated MNI152NLin6 in the file naming structure37. While there are a symmetric and asymmetric version of the MNI152NLin6 T1-weighted image, Neuroparc atlases are registered to the symmetric version, which is used by both FSL 5.031 and new versions of SPM. However, code provided in Neuroparc allows for the registering of any atlas to any reference brain the user chooses.
This image is stored in a GNU-zipped NIfTI file format of a T1-weighted MRI and is available in Neuroparc at three resolutions (1 mm3, 2 mm3, and 4 mm3) for easy use when registering. The naming convention for these files clearly displays their source and resolution as: MNI152NLin6_res-<resolution>_T1w.nii.gz. For example, the format of the resolution input would be “1 × 1 × 1” for the 1 mm3 resolution.
Atlas images and processing
The atlas images compiled in Neuroparc were stored in the form of GNU-zipped NifTi files containing the parcellated atlas. In these files, each region of interest (ROI) within the parcellated image is denoted by a unique integer ranging from 1 to n, where n is the total number of ROIs. Atlases were resampled to the desired voxel resolution through the use of AFNI’s 3dresample30 and then registered to the MNI image of the same resolution using FSL’s flirt function31. The naming convention for the resulting atlas was: <atlas_name>_space-MNI152NLin6_res-<resolution>.nii.gz. The atlas_name field is unique for each atlas image, ideally no more than two words long without a space in between (e.g. Yeo-1738, Princetonvisual36, HarvardOxford27).
Atlas metadata
Using a Python script in Neuroparc, a JSON file containing relevant meta-data was generated for each of the atlases. This file was split into two sections: region-wide and atlas-wide information. The naming convention of the JSON file follows that of the atlas image, with an atlas with 1 mm3 voxel size being named <atlas name>_space-MNI152NLin6_res-1 × 1 × 1 .json.
The term “region-wide” refers to information unique to each region of interest (ROI) in said atlas. This information includes the voxel value for that ROI in the atlas, an anatomical label (if possible), the coordinates of the center of the ROI, and the number of voxels that make up that region. The center and size can be calculated using provided code in the scripts directory of Neuroparc.
Although the label must be specified, this information is not relevant for all atlases. For example, atlases that are generated using algorithmic means, like Slab34, have ROIs that do not strongly correlate to individual anatomical regions. In that case, NULL should be used for the labels of the regions. For ROIs that do have anatomical significance, the naming should follow a hierarchical format in order of largest region to smallest with an underscore in between each name. Modifiers, such as “Superior” or “Medial” can be placed before the anatomical region. An example of this is in the Desikan atlas39, which contains the region with label “L _rostral _anterior _cingulate_cortex”. The main purpose of labeling is to clearly convey the location of an ROI and any anatomical significance it may have. The avoidance of unique abbreviations or terminology that is not widely used helps with the ease of use for individuals new to MRI analysis. Figure 4 shows an example json file.
Optional fields in the region-wide data include description and color. Description can be used to provide more information than the region label if necessary. An example of this use is in the Yeo-7 Networks atlas38. The label for this atlas is in the form ‘7Networks _2’, but the description for that label is the corresponding functional network, ‘Somatomotor’ in this case. The color field must be given in the form [R, G, B] and is only used if the user wants to specify the colors of the regions upon visualization.
Brain-wide data must include the name, description, native coordinate space, and source of the atlas. The name field allows for more elaboration than in the name of the file. The description is more flexible, allowing the creator of an atlas to briefly describe important information for users of their atlas. The intended use case or the method of generation are examples of information provided in this field. Since all atlases in Neuroparc are stored in the same coordinate space, the coordinate space used during the creation of the atlas must be specified.
Finally, the publication detailing the atlas should be included in the source field so users can have a more full understanding of the atlas being used. Optional fields for brain-wide data can all be calculated, including the number of regions, the average volume per region, whether the segmented regions are hierarchical, and if the atlas is symmetrical.
The full description and format of the atlas specification is available within Neuroparc at https://github.com/neurodata/neuroparc/tree/master/atlases/atlas_spec.md.
Dice coefficient
As a way to compare the different atlases to each other, since each has been registered to MNI space, we calculated the Dice Coefficient between atlases. The Dice coefficient is a measure of similarity between two sets40. Specifically, it measures a coincidence index (CI) between two sets, normalized by the size of the sets. Let h be the number of points overlapping in the sets A and B, and a and b are the sizes of their corresponding sets. If the two sets are labelled regions in segmented images, then the Dice coefficient between any pair of regions between the images is given by
where i is the region in image 1 and j is the region in image 2. The result is a similarity matrix, as shown in Fig. 2. Since this map visualizes similarity between two regions in two atlases, the information provided by the Dice map can be used quantify which regions in a given atlas are most similar to regions in another atlas. This method has proven valuable for performing inference with parcellations lacking anatomical annotation, as it allows conclusions realized at the parcel level to be inferred at the anatomical level41.
Adjusted mutual information
Adjusted mutual information is another measure of the similarity of two labelled sets, quantifying how well a particular point can be identified as belonging to a region given another region. It differs from the Dice coefficient in that it tends to be more sensitive to region size and position relative to other measures42.
Similar to the Dice coefficient, Adjusted mutual information is not dependent on a region’s label43. Volumes that share many points are likely to be have a higher mutual information score all else being equal44.
To assure that all atlas comparisons were on the same scale, Neuroparc computes the adjusted mutual information score. Let H(·) denote entropy, N be the number of elements (voxels) in total, and E(MIAB) denote the expected mutual information for sets of size a and b. Here, PA(i) is the probability that a point chosen randomly from the set A will belong to region i45.
where PA,B(a, b) is the probability that a voxel will belong to region a in set A and region b in set B.
where ai is the number of voxels in region i of set A and bj is the number of voxels in region j of set B. \({({a}_{i}+{b}_{j}-N)}^{+}=max(1,{a}_{i}+{b}_{j}-N)\).
as provided in46.
Figure 3 shows the adjusted mutual information between all pairs of atlases. The information provided for this score is atlas-wide, while the Dice score was computed per region to generate a map. The similarity between groups of atlases, such as the various Schaefer atlases, the Yeo liberal atlases, and the DS atlases, is immediately apparent. Recent work highlighting the complementary information provided by disparate parcellations stresses the importance of the availability and ease-of-use of a collection of parcellations from heterogeneous sources41,47,48,49.
Data availability
All atlases and scripts described in this paper are available through a Github repository https://github.com/neurodata/neuroparc. A more extensive repository can be found in the neuroparc OSF registered repository1, which also contains all Dice and AMI figures and adjacency matrix results.
Code availability
Code for processing is publicly available and can be found on GitHub under the scripts folder (https://github.com/neurodata/neuroparc). Examples of useful functions include resampling parcellations to a desired voxel size, the ability to register parcellations to any given reference image, and center calculation for regions of interest for 3D parcellations. Jupyter notebook tutorials are also available for learning how to prepare atlases for being added to Neuroparc. All code is provided under the Apache 2.0 License.
Visualizations are generated using both MIPAV 8.0.2 and FSLeyes 5.0.10 to view the brain volumes in 2D and 3D spaces50,51. Figure 1 can be created using MIPAV triplanar views of each atlases with a striped LUT.
References
Glasser, M. F. et al. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178 (2016).
Fox, M. D. & Greicius, M. Clinical applications of resting state functional connectivity. Front. Systems Neurosci. 4, 19 (2018).
Eickhoff, S. B., Yeo, B. T. & Benon, S. Imaging-based parcellations of the human brain. Nature Reviews Neuroscience 19, 672–686 (2018).
Hagmann, P. et al. Mapping human whole-brain structural networks with diffusion mri. PloS one 2, e597 (2007).
Zalesky, A. et al. Whole-brain anatomical networks: does the choice of nodes matter? Neuroimage 50, 970–983 (2010).
Bellec, P., Rosa-Neto, P., Lyttelton, O. C., Benali, H. & Evans, A. C. Multi-level bootstrap analysis of stable clusters in resting-state fmri. Neuroimage 51, 1126–1139 (2010).
Craddock, R. C., James, G. A., Holtzheimer, P. E. III, Hu, X. P. & Hayberg, H. S. A whole brain fmri atlas generated via spatially constrained spectral clustering. Human brain mapping 33, 1914–1928 (2012).
Nikolaidis, A. et al. Bagging improves reproducibility of functional parcellation. Neuroimage 214, 343–392 (2020).
Thirion, B., Varoquaux, G., Dohmatob, E. & Poline, J. B. Which fmri clustering gives good brain parcellations? Front. Neurosci. 8, 167 (2014).
Beckmann, C. F., Mackay, C. E., Filippini, N. & Smith, S. M. Group comparison of resting-state fmri data using multi-subject ica and dual regression. Neuroimage 47, S148 (2009).
Varoquaux, G., Gramfort, A., Pedregosa, F., Michel, V. & Thirion, B. Multi-subject dictionary learning to segment an atlas of brain spontaneous activity. Biennial International Conference on Information Processing in Medical Imaging 6801, 562–573, https://doi.org/10.1007/978-3-642-22092-0_46 (2011).
Cohen, A. L. et al. Defining functional areas in individual human brains using resting functional connectivity mri. Neuroimage 41, 45–57 (2008).
Gordon, E. M. et al. Generation and evaluation of cortical area parcellation from resting-state correlations. Cerebral cortex 26, 288–303 (2014).
Nelson, S. M. et al. A parcellation scheme for human left lateral parietal cortex. Neuron 67, 156–170 (2010).
Wig, G. S. et al. Parcellating an individual subject’s cortical and subcortical brain structures using snowball sampling of resting-state correlations. Cerebral cortex 24, 2036–2054 (2013).
Xu, T. et al. Assessing variations in a real organization for the intrinsic brain: from fingerprints to reliability. Cerebral Cortex 26, 4192–4211 (2016).
Margulies, D. S. et al. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proceedings of the National Academy of Sciences 113, 12574–12579 (2016).
Dosenbach, N. U. et al. Prediction of individual brain maturity using fmri. Science 329, 1358–1361 (2010).
Liem, F. et al. Predicting brain-age from multimodal imaging data captures cognitive impairment. Neuroimage 148, 179–188 (2017).
Finn, E. S. et al. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nature neuroscience 18, 1664 (2015).
Shehzad, Z. et al. A multivariate distance-based analytic framework for connectome-wide association studies. Neuroimage 93, 74–94 (2014).
Abraham, A. et al. Deriving reproducible biomarkers from multi-site resting-state data: an autism-based example. Neuroimage 147, 736–745 (2017).
Greene, D. J. et al. Multivariate pattern classification of pediatric tourette syndrome using functional connectivity mri. Developmental science 19, 581–598 (2016).
Hong, S. J., Valk, S. L., Di Martino, A., Milham, M. P. & Bernhardt, B. C. Multidimensional neuroanatomical subtyping of autism spectrum disorder. Cerebral Cortex 28, 3578–3588 (2017).
Arslan, S. et al. Human brain mapping: A systematic comparison of parcellation methods for the human cerebral cortex. Neuroimage 170, 5–30 (2018).
Abraham, A. et al. Machine learning for neuroimaging with scikit-learn. Front. Neuroinfo. 8, 14 (2014).
Markis, N. et al. Decreased volume of left and total anterior insular lobule in schizophrenia. Schizophrenia Research 83, 155–171, https://doi.org/10.1016/j.schres.2005.11.020 (2006).
Tzourio-Mazoyer, N. et al. Automated anatomical labeling of activations in spm using a macroscopic anatomical parcellation of the mri single-subject brain. Neuroimage 15, 273–289, https://doi.org/10.1006/nimg.2001.0978 (2002).
Neurodata. Neuroparc Open Science Framework https://doi.org/10.17605/OSF.IO/DVXW3 (2020).
Cox, R. W. Afni: Software for analysis and visualisation of functional magnetic resonance neuroimages. Compt. Biomed. Res. 29, 162–173, https://doi.org/10.1006/cbmr.1996.0014 (1996).
Jenkinson, M. & Smith, S. A global optimisation method for robust affine registration of brain images. Medical Image Analysis 5, 143–156 (2001).
Schaefer, A. et al. Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cerebral Cortex 28, 3095–3114 (2018).
Mhembere, D. et al. Computing scalable multivariate Local invariants of large (brain-) fraphs. 2013 IEEE Global Conference on Signal and Information Processing, 297–300, https://doi.org/10.1109/GlobalSIP.2013.6736874 (2013).
Sripada, C. S., Kessler, D. & Angstadt, M. Lag in maturation of the brain’s intrinsic functional architecture in attention-deficit/hyperactivity disorder. Proceedings of the National Academy of Sciences 111, 14259–14264, https://doi.org/10.1073/pnas.1407787111 (2014).
Hua, K. et al. Tract probability maps in stereotaxic spaces; analyses of white matter anatomy and tract-specific quantification. Neuroimage 39, 336–347 (2007).
Wang, L., Mruczek, R. E. B., Arcaro, M. J. & Kastner, S. Probabilistic maps of visual topography in human cortex. Cerebral Cortex 25, 3911–3931 (2015).
Chau, W. & McIntosh, A. R. The talairach coordinates of a point in the MNI space: how to interpret it. Neuroimage 25, 408–416, https://doi.org/10.1016/j.neuroimage.2004.12.007 (2004).
Yeo, B. T. T. et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of Neurophysiology 106, 1125–1165 (2011).
Desikan, R. S. et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980, https://doi.org/10.1016/j.neuroimage.2006.01.021 (2006).
Dice, L. R. Measures of the amount of ecologic association between species. Ecology 26, 297–302 (1945).
Priebe, C. E. et al. On a two-truths phenomenon in spectral graph clustering. Proc. Natl. Acad. Sci. USA 116, 5995–6000 (2019).
Zou, K. H., Wells, W. M., Kikinis, R. & Warfield, S. K. Three validation metrics for automated probabilistic image segmentation of brain tumors. Stat Med 23, 1259–1282 (2004).
Lin, D. An information-theoretic definition of similarity. Proceedings of the Fifteenth International Conference on Machine Learning 296–304 (1998).
Mitchell, H. B. Image similarity measures. In Image Fusion., 167–185 (2010).
Tateoka, K. Assessment of similarity measures for accurate deformable image registration. Journal of Nuclear Medicine & Radiation Therapy 3, https://doi.org/10.4172/2155-9619.1000137 (2012).
Vinh, N. X., Epps, J. & Bailey, J. Information theoretic measures for clustering comparison: Variants, properties, normalization and correction for chance. Journal of Machine Learning Research 11, 2837–2853 (2010).
Wu, Z., Xu, D., Potter, T. & Zhang, Y. & Alzheimer’s Disease Neuroimaging Initiative. Effects of brain parcellation on the characterization of topological deterioration in Alzheimer’s disease. Front. Aging Neurosci. 11, 113 (2019).
Messe, A. Parcellation influence on the connectivity-based structure-function relationship in the human brain. Hum. Brain Mapp. 41, 1167–1180 (2020).
Wang, J. et al. Parcellation-dependent small-world brain functional networks: a resting-state fMRI study. Hum. Brain Mapp. 30, 1511–1523 (2009).
McAuliffe, M. J. et al. Medical image processing, analysis and visualization in clinical research. In Proceedings 14th IEEE Symposium on Computer-Based Medical Systems. CBMS 2001, 381–386, https://doi.org/10.1109/CBMS.2001.9411749 (2001).
McCarthy, P. Source code for: FSLeyes Zenodo https://doi.org/10.5281/zenodo.1470761 (2020).
Brodmann, K. Vergleichende Lokalisationslehre der Groçhirnrinde: in ihren Prinzipiendargestellt auf Grund des Zellenbaues. (1909).
Giavasis, S. et al. Fcp-Indi/C-Pac: Cpac Version 1.0.0 Beta. Zenodo https://doi.org/10.5281/zenodo.164638 (2016).
Talairach, J. & Szikla, G. Application of stereotactic concepts to the surgery of epilepsy. Acta Neurochirurgica. Supplementum 30, 35–54 (1980).
Joliot, M. et al. AICHA: An atlas of intrinsic connectivity of homotopic areas. Neurosci Methods. 254, 46–59 (2015).
Fischl, B. et al. Automatically parcellating the human cerebral cortex. Cereb Cortex. 14, 11–22 (2004).
Kevin, A. & Tourville, J. 101 labeled brain images and a consistent human cortical labeling protocol. Front. Neurosci. 6, 171 (2012).
Ioannis, S. et al. Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. NeuroImage 40, 672–684 (2008).
Simon, B. et al. A new SPM toolbox for combining probabilistic cytoarchitectonic maps and functional imaging data. NeuroImage 25, 1053–8119 (2005).
Landman, B. et al. MICCAI 2012 workshop on multi-atlas labeling. CreateSpace 2 (2012).
Acknowledgements
We would like to acknowledge generous support from National Science Foundation (NSF) under NSF Award Number EEC-1707298. Reasearch was partially supported by funding from Microsoft Research.
Author information
Authors and Affiliations
Contributions
R.L., E.W.B., P.M., G.A., D.A.P. and J.T.V. prepared the data source and performed the comparative data analysis. S.R., P.F., A.L. and A.N. performed the technical validation. All authors prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lawrence, R.M., Bridgeford, E.W., Myers, P.E. et al. Standardizing human brain parcellations. Sci Data 8, 78 (2021). https://doi.org/10.1038/s41597-021-00849-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-00849-3
This article is cited by
-
Assessment of brain cancer atlas maps with multimodal imaging features
Journal of Translational Medicine (2023)
-
Should one go for individual- or group-level brain parcellations? A deep-phenotyping benchmark
Brain Structure and Function (2023)