Abstract
DNA methylation is an important epigenetic modification1,2,3. Ten-eleven translocation (TET) proteins are involved in DNA demethylation through iteratively oxidizing 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC)4,5,6,7,8. Here we show that human TET1 and TET2 are more active on 5mC-DNA than 5hmC/5fC-DNA substrates. We determine the crystal structures of TET2–5hmC-DNA and TET2–5fC-DNA complexes at 1.80 Å and 1.97 Å resolution, respectively. The cytosine portion of 5hmC/5fC is specifically recognized by TET2 in a manner similar to that of 5mC in the TET2–5mC-DNA structure9, and the pyrimidine base of 5mC/5hmC/5fC adopts an almost identical conformation within the catalytic cavity. However, the hydroxyl group of 5hmC and carbonyl group of 5fC face towards the opposite direction because the hydroxymethyl group of 5hmC and formyl group of 5fC adopt restrained conformations through forming hydrogen bonds with the 1-carboxylate of NOG and N4 exocyclic nitrogen of cytosine, respectively. Biochemical analyses indicate that the substrate preference of TET2 results from the different efficiencies of hydrogen abstraction in TET2-mediated oxidation. The restrained conformation of 5hmC and 5fC within the catalytic cavity may prevent their abstractable hydrogen(s) adopting a favourable orientation for hydrogen abstraction and thus result in low catalytic efficiency. Our studies demonstrate that the substrate preference of TET2 results from the intrinsic value of its substrates at their 5mC derivative groups and suggest that 5hmC is relatively stable and less prone to further oxidation by TET proteins. Therefore, TET proteins are evolutionarily tuned to be less reactive towards 5hmC and facilitate the generation of 5hmC as a potentially stable mark for regulatory functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002)
Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet. 33 (Suppl.), 245–254 (2003)
Smith, Z. D. & Meissner, A. DNA methylation: roles in mammalian development. Nature Rev. Genet. 14, 204–220 (2013)
Guo, F. et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–458 (2014)
He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011)
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011)
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009)
Wang, L. et al. Programming and inheritance of parental DNA methylomes in mammals. Cell 157, 979–991 (2014)
Hu, L. et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell 155, 1545–1555 (2013)
Globisch, D. et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 5, e15367 (2010)
Mellén, M., Ayata, P., Dewell, S., Kriaucionis, S. & Heintz, N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430 (2012)
Pfaffeneder, T. et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew. Chem. Int. Edn Engl. 50, 7008–7012 (2011)
Song, C. X. et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nature Biotechnol. 29, 68–72 (2011)
Shen, L. et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153, 692–706 (2013)
Hashimoto, H. et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature 506, 391–395 (2014)
Song, C. X. et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 153, 678–691 (2013)
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012)
Liu, H., Llano, J. & Gauld, J. W. A DFT study of nucleobase dealkylation by the DNA repair enzyme AlkB. J. Phys. Chem. B 113, 4887–4898 (2009)
Yang, C. G. et al. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 452, 961–965 (2008)
Aik, W., McDonough, M. A., Thalhammer, A., Chowdhury, R. & Schofield, C. J. Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases. Curr. Opin. Struct. Biol. 22, 691–700 (2012)
Price, J. C., Barr, E. W., Glass, T. E., Krebs, C. & Bollinger, J. M., Jr. Evidence for hydrogen abstraction from C1 of taurine by the high-spin Fe(IV) intermediate detected during oxygen activation by taurine:alpha-ketoglutarate dioxygenase (TauD). J. Am. Chem. Soc. 125, 13008–13009 (2003)
Kaizer, J. et al. Nonheme FeIVO complexes that can oxidize the C–H bonds of cyclohexane at room temperature. J. Am. Chem. Soc. 126, 472–473 (2004)
de Visser, S. P., Kumar, D., Cohen, S., Shacham, R. & Shaik, S. A predictive pattern of computed barriers for C–H hydroxylation by compound I of cytochrome P450. J. Am. Chem. Soc. 126, 8362–8363 (2004)
Münzel, M. et al. Improved synthesis and mutagenicity of oligonucleotides containing 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine. Chemistry 17, 13782–13788 (2011)
Wondrack, L. M., Hsu, C. A. & Abbott, M. T. Thymine 7-hydroxylase and pyrimidine deoxyribonucleoside 2′ -hydroxylase activities in Rhodotorula glutinis . J. Biol. Chem. 253, 6511–6515 (1978)
Baggaley, K. H., Brown, A. G. & Schofield, C. J. Chemistry and biosynthesis of clavulanic acid and other clavams. Nat. Prod. Rep. 14, 309–333 (1997)
Fu, Y. et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nature Commun. 4, 1798 (2013)
Blaschke, K. et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226 (2013)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
DeLano, W. L. The PyMOL molecular graphics system (DeLano Scientific, 2002)
Frisch, M. et al. Gaussian 09, Revision A.02 (Gaussian, 2009).
Montgomery, J. A. Jr, Frisch, M. J., Ochterski, J. W. & Petersson, G. A. A complete basis set model chemistry. VII. Use of the minimum population localization method. J. Chem. Phys. 112, 6532–6542 (2000)
Curtiss, L. A., Redfern, P. C. & Raghavachari, K. Gaussian-4 theory. J. Chem. Phys. 126, 084108 (2007)
Baboul, A. G., Curtiss, L. A., Redfern, P. C. & Raghavachari, K. Gaussian-3 theory using density functional geometries and zero-point energies. J. Chem. Phys. 110, 7650 (1999)
Cossi, M., Rega, N., Scalmani, G. & Barone, V. Energies, structures, and electronic properties of molecules in solution with the C-PCM solvation model. J. Comput. Chem. 24, 669–681 (2003)
Case, D. A. et al. Amber 11 (Univ. California, 2010)
Pellegrini, E. & Field, M. J. A generalized-Born solvation model for macromolecular hybrid-potential calculations. J. Phys. Chem. A 106, 1316–1326 (2002)
Roberts, R. J. & Cheng, X. Base flipping. Annu. Rev. Biochem. 67, 181–198 (1998)
Luo, Y. R. Comprehensive Handbook of Chemical Bond Energies Ch. 3 (CRC, 2007)
Acknowledgements
We thank the staff of beamlines BL17U and BL19U at Shanghai Synchrotron Radiation Facility for assistance in data collection, and staff of the Biomedical Core Facility, Fudan University, for their help with mass spectrometry. The computation resources were supported by the Computer Network Information Center, Chinese Academy of Sciences and Shanghai Supercomputing Center. This work was supported by grants from the National Basic Research Program of China (2011CB965300), the National Science & Technology Major Project ‘Key New Drug Creation and Manufacturing Program’ of China (2014ZX09507-002), the National Natural Science Foundation of China (U1432242, 31425008, 91419301, 91313000, 31270779, 21210003), the Basic Research Project of Shanghai Science and Technology Commission (12JC1402700), the Program of Shanghai Subject Chief Scientist (14XD1400500), the Hi-Tech Research and Development Program of China (2012AA020302 and 2012AA01A305) and the Chinese Academy of Sciences (XDA01040305).
Author information
Authors and Affiliations
Contributions
L.H., J.L., J.C., C.L. and Y.X. designed the experiments. L.H., J.C., Z.L., Q.R., W.G., M.L., C.S. and X.Y. purified protein. L.H. performed crystallization and all enzymatic assays. L.H., J.C., H.H., Z.L. and J.L. collected the data and determined the crystal structure. J.C. and L.H. performed the fluorescence polarization and SPR assays. L.H., J.C., W.L. and X.T. performed stopped-flow analyses. L.Z. and P.Y. designed and performed LC–MS/MS and analysed the data. J.L., H.J., C.L., D.F. and Q.C. designed and performed computational studies and analysed the data. C.H. and Y.W. provided standard for LC–MS/MS analyses. L.H., J.L., C.L. and Y.X. analysed the data and wrote the manuscript. Y.X. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
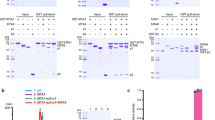
Extended Data Figure 1 Steady-state kinetic analyses for TET-mediated oxidation on 5mC/5hmC/5fC-DNA substrates.
a, Protein purification of human TET1 catalytic domain. Representative gel-filtration profile of human TET1 (residues 1418–2136) is shown. The peak position is about 13 ml, which corresponds to the monomer of TET1 with molecular mass of about 79 kilodaltons. The peak fractions were subjected to SDS–polyacrylamide gel electrophoresis and stained by Coomassie blue. The column used for gel filtration was Superdex 200 (GE Healthcare, 10/300 GL). b, Standard curves for 5mC, 5hmC, 5fC, 5caC, [2H]35mC, [2H]35hmC and guanine for quantification in LC–MS/MS. Good linearity was obtained for the range of guanine and various cytosine derivatives as indicated. The level of guanine (equal to total cytosine and its derivatives) was detected. Note that three standard curves were generated for 5mC/5hmC/5fC in low/mid/high concentrations, respectively. Two standard curves were generated for 5caC and [2H]35mC in mid/low-concentration. c–e, Reaction progress curves of substrate and fraction products versus incubation time (2.5 min). Initial rates (nanomoles per second) for product generation were measured using various concentrations of dsDNA substrate and TET2. The reactions were conducted using 58-bp dsDNA (D5, one central 5mC/5hmC/5fCpG site) as substrate. Quantification was calculated from two independent assays; error bars, s.d. for duplicate experiments. f–h, To avoid generation of multiple products, we optimized protein concentration so that only one product was detected for all the reactions under our experimental conditions. The product generations are shown for the reactions with the highest substrate concentration for the longest time.
Extended Data Figure 2 Similar substrate preference of TET2 towards DNA with different lengths and sequences.
a–e, Enzymatic activity of TET2 was measured on 5mC/5hmC/5fC-DNA substrates (one central 5mC/5hmC/5fCpG site) of different lengths (a–c) or sequences (b, d, e). dsDNA substrates of 26 bp, 58 bp and 100 bp were used for reactions to test the effect of DNA length on substrate preference of TET2 (a–c). AT- and CG-rich dsDNA substrates of 58 bp were used for reactions to compare the effect of DNA sequence on substrate preference of TET2 (b, d, e). TET2 (1 μM) was used for all reactions. Quantification was calculated from three independent assays; error bars, s.d. for triplicate experiments. TET2 showed similar substrate preference (higher activity on 5mC-DNA than on 5hmC/5fC-DNA) for all DNA substrates tested. In addition, TET2 has no sequence preference apart from CpG dinucleotide. Notably, TET2 showed higher activity on shorter DNA substrate under our experimental conditions. It is not surprising because the enzyme should be able to find one CpG site easier on shorter DNA substrate (compared with longer DNA). f–h, Effects of substrate/product on enzymatic activities of TET2. f, Enzymatic activities of 1 μM TET2 for 26-bp 5mC-DNA in the presence of 26-bp 5C/5hmC-DNA. g, Enzymatic activities of 1 μM TET2 for 26-bp 5hmC-DNA in the presence of 26-bp 5C/5fC-DNA. h, Enzymatic activities of 1 μM TET2 for 26-bp 5fC-DNA in the presence of 26-bp 5C/5caC-DNA. The presence of CpG-DNA or different substrate/products has negligible impact on TET2 activity.
Extended Data Figure 3 Fluorescence polarization measurements of DNA-binding affinities of TET2 in different conditions.
a–c, Fluorescence polarization measurements of substrate DNA-binding affinities of TET2 (10 mM HEPES pH 7.0, 100 mM NaCl) in the presence of 100 μM Mn2+ and 1 mM 2-OG (a), 100 μM Fe2+ and 1 mM NOG (b) and 100 μM Fe2+ and 1 mM succinate (c). No significant difference was observed for the DNA-binding affinity of TET2 for different substrate/product under conditions mimicking oxidation or after oxidation.
Extended Data Figure 4 Structural comparison of TET2–5mC-DNA, TET2–5hmC-DNA and TET2–5fC-DNA complexes.
a–c, Structural comparison of the three complexes. The individual structures of the three complexes are shown on the left panel and the superimposed structures are shown on the right. The structures are shown in ribbon representations. TET2–5hmC-DNA and TET2–5fC-DNA are coloured as in Fig. 3b, and TET2–5mC-DNA is coloured in grey. The colour scheme is indicated. Stick representations show 5mC and 5hmC in two structures. d, Close-up view for the comparison of 5mC, 5hmC and 5fC in the three structures. Note that the cytosine portions of the three bases adopt almost identical conformations within the catalytic cavity. e–g, Close-up views of the catalytic DSBH core of the three structures, shown in ribbon representation with critical residues indicated in stick representations. The nitrogen and oxygen atoms are coloured in blue and red, respectively. Hydrogen bonds and Fe(II) coordination are indicated as dashed lines. Fe(II) and crystallographic water molecules are shown as red and green balls, respectively. The cytosine of 5hmC is specifically recognized by TET2 in a similar manner to that of 5mC in the TET2–5mC-DNA structure. An additional hydrogen bond between the hydroxymethyl group of 5hmC and NOG was observed in the TET2–5hmC-DNA structure. The additional hydrogen bond may not be strong enough to affect the binding affinity between TET2 and DNA because the interaction is mediated by extensive hydrogen bonds and hydrophobic interactions (Extended Data Fig. 5).
Extended Data Figure 5 Recognition of CpG dinucleotide by TET2.
a, Two different views of the interaction between the G7:hmC7′ base pair and TET2 for the specific recognition of CpG dinucleotide in TET2–5hmC-DNA. Water-mediated hydrogen bonds are formed between base hmC7′ of DNA and residues Y1295 and R1302 of TET2. The 2Fobserved − Fcalculated simulated annealing omit maps for residues involved in CpG recognition outside catalytic cavity are shown. The maps were calculated at 1.80 Å and contoured at 1.0σ. b, Two different views of the interaction between the G7:C7′ base pair and TET2 for the specific recognition of CpG dinucleotide in TET2–5fC-DNA. The 2Fobserved − Fcalculated simulated annealing omit maps were calculated at 1.97 Å and contoured at 1.0σ. c, d, Close-up views of the interactions between TET2 and 5hmC-DNA (c) and TET2 and 5fC-DNA (d). Critical bases of DNA and residues of TET2 for the interactions are shown in stick representations. Hydrogen bonds are indicated as dashed lines. Water molecule is shown as a green ball. The nitrogen, oxygen and phosphorus atoms are coloured in blue, red and orange, respectively. The 2Fobserved − Fcalculated simulated annealing omit maps for residues involved in DNA interaction are shown with these residues omitted from the calculation. Most residues are well covered by the electron density, indicating that these residues were built correctly in the structural model. e, f, Representation of intermolecular contacts between TET2 and 5hmC-DNA (e) and 5fC-DNA (f).
Extended Data Figure 6 Kinetic traces from the reactions of TET2·Fe(II)·2-OG.
Traces from the reactions in the absence (a) and presence (b) of unmethylated CpG-DNA or random DNA (c). The formation of catalytic intermediate for each reaction was monitored by stopped-flow absorption at 318 nm. No decay of catalytic intermediate was observed for any of the measurements. These analyses serve as a negative control for the assays shown in Fig. 4c.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-7. (PDF 139 kb)
Rights and permissions
About this article
Cite this article
Hu, L., Lu, J., Cheng, J. et al. Structural insight into substrate preference for TET-mediated oxidation. Nature 527, 118–122 (2015). https://doi.org/10.1038/nature15713
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature15713
This article is cited by
-
The Role of 5-Hydroxymethylcytosine as a Potential Epigenetic Biomarker in a Large Series of Thyroid Neoplasms
Endocrine Pathology (2024)
-
Alterations of the expression of TET2 and DNA 5-hmC predict poor prognosis in Myelodysplastic Neoplasms
BMC Cancer (2023)
-
TET3 as a non-invasive screening tool for the detection of fibrosis in patients with chronic liver disease
Scientific Reports (2023)
-
Flanking sequences influence the activity of TET1 and TET2 methylcytosine dioxygenases and affect genomic 5hmC patterns
Communications Biology (2022)
-
Two families with TET3-related disorder showing neurodevelopmental delay with craniofacial dysmorphisms
Journal of Human Genetics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.