Abstract
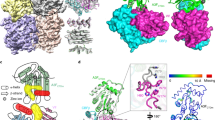
The human immunodeficiency virus (HIV)-1 protein Vif has a central role in the neutralization of host innate defences by hijacking cellular proteasomal degradation pathways to subvert the antiviral activity of host restriction factors1,2,3,4,5,6; however, the underlying mechanism by which Vif achieves this remains unclear. Here we report a crystal structure of the Vif–CBF-β–CUL5–ELOB–ELOC complex. The structure reveals that Vif, by means of two domains, organizes formation of the pentameric complex by interacting with CBF-β, CUL5 and ELOC. The larger domain (α/β domain) of Vif binds to the same side of CBF-β as RUNX1, indicating that Vif and RUNX1 are exclusive for CBF-β binding. Interactions of the smaller domain (α-domain) of Vif with ELOC and CUL5 are cooperative and mimic those of SOCS2 with the latter two proteins. A unique zinc-finger motif of Vif, which is located between the two Vif domains, makes no contacts with the other proteins but stabilizes the conformation of the α-domain, which may be important for Vif–CUL5 interaction. Together, our data reveal the structural basis for Vif hijacking of the CBF-β and CUL5 E3 ligase complex, laying a foundation for rational design of novel anti-HIV drugs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mariani, R. et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31 (2003)
Sheehy, A. M., Gaddis, N. C. & Malim, M. H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nature Med. 9, 1404–1407 (2003)
Yu, X. et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060 (2003)
Stopak, K., de Noronha, C., Yonemoto, W. & Greene, W. C. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12, 591–601 (2003)
Marin, M., Rose, K. M., Kozak, S. L. & Kabat, D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nature Med. 9, 1398–1403 (2003)
Conticello, S. G., Harris, R. S. & Neuberger, M. S. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13, 2009–2013 (2003)
Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002)
Mangeat, B. et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003)
Harris, R. S. et al. DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 (2003)
Zhang, H. et al. The cytidinedeaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424, 94–98 (2003)
Lecossier, D., Bouchonnet, F., Clavel, F. & Hance, A. J. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300, 1112 (2003)
Stanley, B. J. et al. Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J. Virol. 82, 8656–8663 (2008)
Luo, K. et al. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5–E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl Acad. Sci. USA 102, 11444–11449 (2005)
Zhang, W., Du, J., Evans, S. L., Yu, Y. & Yu, X. F. T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481, 376–379 (2011)
Jager, S. et al. Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection. Nature 481, 371–375 (2011)
Kim, Y. K. et al. Structural basis of intersubunit recognition in elongin BC-cullin 5-SOCS box ubiquitin-protein ligase complexes. Acta Crystallogr. D 69, 1587–1597 (2013)
Zheng, N. et al. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709 (2002)
Stebbins, C. E., Kaelin, W. G., Jr & Pavletich, N. P. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science 284, 455–461 (1999)
Bullock, A. N., Debreczeni, J. E., Edwards, A. M., Sundstrom, M. & Knapp, S. Crystal structure of the SOCS2-elongin C-elongin B complex defines a prototypical SOCS box ubiquitin ligase. Proc. Natl Acad. Sci. USA 103, 7637–7642 (2006)
Zhou, X., Evans, S. L., Han, X., Liu, Y. & Yu, X. F. Characterization of the interaction of full-length HIV-1 Vif protein with its key regulator CBFβ and CRL5 E3 ubiquitin ligase components. PLoS ONE 7, e33495 (2012)
Taniuchi, I. et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002)
Warren, A. J., Bravo, J., Williams, R. L. & Rabbitts, T. H. Structural basis for the heterodimeric interaction between the acute leukaemia-associated transcription factors AML1 and CBFβ. EMBO J. 19, 3004–3015 (2000)
Kim, D. Y. et al. CBF-β stabilizes HIV Vif to counteract APOBEC3 at the expense of RUNX1 target gene expression. Mol. Cell 49, 632–644 (2013)
Hultquist, J. F., McDougle, R. M., Anderson, B. D. & Harris, R. S. HIV type 1 viral infectivity factor and the RUNX transcription factors interact with core binding factor β on genetically distinct surfaces. AIDS Res. Hum. Retroviruses 28, 1543–1551 (2012)
Xiao, Z. et al. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology 349, 290–299 (2006)
Zimmerman, E. S., Schulman, B. A. & Zheng, N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 20, 714–721 (2010)
Chen, G., He, Z., Wang, T., Xu, R. & Yu, X. F. A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J. Virol. 83, 8674–8682 (2009)
Russell, R. A. & Pathak, V. K. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81, 8201–8210 (2007)
Zhang, H., Pomerantz, R. J., Dornadula, G. & Sun, Y. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J. Virol. 74, 8252–8261 (2000)
He, Z., Zhang, W., Chen, G., Xu, R. & Yu, X. F. Characterization of conserved motifs in HIV-1Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381, 1000–1011 (2008)
Otwinowski, Z. M. W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Muniz, J. R. C. et al. Molecular architecture of the ankyrin SOCS box family of Cul5-dependent E3 ubiquitin ligases. J. Mol. Biol. 425, 3166–3177 (2013)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
DeLano, W. L. PyMOL Molecular Viewer (http://www.pymol.org) (2002)
Acknowledgements
We thank F. Yu and J. He at Shanghai Synchrotron Radiation Facility (SSRF) for help with data collection. We thank J. Chai for critical reading of the manuscript. This research was funded by the National Natural Science Foundation of China grant 31300605 and the Program for New Century Excellent Talents in University grant AUGA5710060713 to Z.H.; Ministry of Science and Technology of China grants 2012CB911101 and 2011CB910502, and the National Natural Science Foundation of China grants 31030020 and 31170679 to M.Y.
Author information
Authors and Affiliations
Contributions
Z.H. designed the experiments. Y.G., L.D. and Z.H. performed the bulk of the experiments. Data were analysed by Z.H., Y.G., L.D., X.Q.; Y.W., B.Z., H.L., Y.Y., Y.Z. and M.Y. contributed to some experiments and discussions. Z.H., Y.G., L.D. and X.Q. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
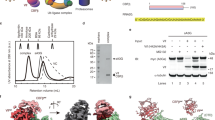
Extended Data Figure 1 Purification of reconstituted Vif–CBF-β–nCUL5–ELOB–ELOC complex protein.
a, Formation of the Vif–CBF-β–nCUL5–ELOB–ELOC pentameric complex in gel filtration. SDS–PAGE and Coomassie blue staining of Vif–CBF-β–nCUL5–ELOB–ELOC complex of the peak fractions from gel filtration. MM, molecular mass marker. b, Limited proteolysis analysis of Vif pentameric complex. The purified complex protein of Vif–CBF-β–nCUL5–ELOB–ELOC was treated with elastase (at a molar ratio of 1:1,000) at 4 °C for 72 h. The elastase-treated hetropentameric protein was visualized by Coomassie blue staining following SDS–PAGE.
Extended Data Figure 2 The density map of a representative region of Vif.
a, The simulated annealing composite omit map of a representative region of Vif. An Fo − Fc omit map is contoured at 3σ in green, and a 2Fo − Fc omit map in blue contoured at 1.3σ. b, The electron density of Zn2+ in Vif. The Zn2+-binding site in Vif is shown. 2Fo − Fc electron density map (blue mesh, contoured at 1.3σ) at the Zn2+-binding site. Four residues involved in Zn2+ interaction are labelled.
Extended Data Figure 3 Structural comparison of Vif–ELOB–ELOC with Vif peptide-bound ELOB–ELOC.
Structural comparison of Vif–ELOB–ELOC with Vif (residues 139–176, green)–ELOB (purple)–ELOC (cyan) (Protein Data Bank accession 3DCG)12. The side chains of 161-PPLPS-165 of Vif are labelled and shown in grey.
Extended Data Figure 4 Structural comparison of Vif–CUL5–ELOC and CUL1–SKP1–SKP2.
a, Interface of CUL5–ELOC. The side chains from CUL5 are labelled and shown in orange, and those from ELOC are coloured in hot pink. Red dashed lines represent hydrogen bonds. b, Interface of CUL1–SKP1. The side chains of CUL1 and SKP1 are labelled and shown in orange and slate, respectively. Residues involved in interactions are numbered and hydrogen bonds are shown as red dashed lines. c, Mutagenesis analyses of the nCUL5–ELOC interface. Wild-type or mutant GST-fused nCUL5 proteins were first bound to glutathione-sepharose beads and incubated with wild-type or various ELOC mutant Vif–CBF-β–ELOB–ELOC complex proteins as indicated. After extensive washing, the bound proteins were visualized by Coomassie blue staining following SDS–PAGE. d, Structural comparison of Vif–ELOC–CUL5 and SKP2 (grey)–SKP1 (blue)–CUL1 (green) (Protein Data Bank 1LDK)17; the interfaces of the two complexes are highlighted in red frames, and secondary structural elements are labelled.
Extended Data Figure 5 CBF-β–nCUL5–ELOB–ELOC bind to the hydrophobic surfaces of Vif, leaving the positively charged patches solvent-exposed.
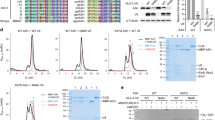
a, Sequence alignment of HIV/SIV Vif proteins. Conserved and similar residues are highlighted with red and yellow shading, respectively. Residues involved in E3 ligase complex and APOBEC3 proteins interaction are indicated with slate solid dots at the bottom. Conserved motifs of zinc-finger region and BC-box motif are indicated in the sequence alignment. b, Two views of the electrostatic surfaces of CBF-β–nCUL5–ELOC bound Vif are presented. CBF-β–nCUL5–ELOC complex binds to the hydrophobic surfaces of Vif (right). Positively charged patches on Vif (left) are solvent-exposed. White, blue and red indicate neutral, positive and negative surfaces, respectively. c, The largely solvent-exposed surface of Vif is shown in ribbon representation and electrostatic surface. The side chains of relevant residues from Vif involved in the interaction with APOBEC3G alone, APOBEC3F alone and both of them are shown in stick representation and are coloured in green, cyan and yellow, respectively.
Extended Data Figure 6 The interaction between the C terminus of CBF-β and Vif.
a, The interface of Vif and helix H5 of CBF-β. Side chains from Vif involved in the interaction are labelled and shown in slate, and those from CBF-β are labelled and shown in yellow. b, C-terminus-truncated CBF-β is inefficient to assemble the Vif–CBF-β–nCUL5–ELOB–ELOC complex. Complexes of 6×His–Vif–CBF-β (1–170)–ELOB–ELOC and 6×His–Vif–CBF-β (1–140)–ELOB–ELOC were purified using Ni-NTA and then incubated with purified nCUL5 (input) before subjecting to size-exclusion chromatography (Superdex 200, Pharmacia). Shown on the top is superposition of the gel filtration chromatograms of two complexes containing CBF-β (1–170) (red) and CBF-β (1–140) (blue). The horizontal axis represents elution volume (volume unit, ml). Middle and bottom: Coomassie blue staining of the peak fractions of two complexes shown on the top after SDS–PAGE.
Extended Data Figure 7 Mutagenesis analysis of residues involved in Vif–ELOC interaction.
Vif–ELOC interaction pull-down assay. Wild-type GST–ELOC–ELOB immobilized on GS4B resin was incubated with wild-type or point mutant Vif–CBF-β proteins, washed and resolved by SDS–PAGE (left panel); wild-type 6×His–Vif–CBF-β protein immobilized on Ni+ resin was incubated with wild-type or point mutant GST–ELOC–ELOB proteins, washed and resolved by SDS–PAGE (right panel).
Extended Data Figure 8 Vif Phe 115 is located in a highly hydrophobic environment of Vif.
The detailed interactions around the residue of Phe 115 of Vif. Vif Phe 115 is surrounded with a hydrophobic environment. The side chains of relevant residues are shown in stick representation and labelled.
Extended Data Figure 9 Vif demonstrates higher binding affinity for nCUL5 than nCUL2 for Vif–CBF-β–nCUL5–ELOB–ELOC complex assembly.
6×His–Vif–CBF-β–ELOB–ELOC was first bound to Ni-NTA beads and then incubated with purified nCUL2 and nCUL5 as indicated. After extensive washing, the bound proteins were visualized by Coomassie blue staining after SDS–PAGE.
Rights and permissions
About this article
Cite this article
Guo, Y., Dong, L., Qiu, X. et al. Structural basis for hijacking CBF-β and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505, 229–233 (2014). https://doi.org/10.1038/nature12884
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12884
This article is cited by
-
Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H
Nature Communications (2023)
-
HIV-1 Vif suppresses antiviral immunity by targeting STING
Cellular & Molecular Immunology (2022)
-
Inhibition of base editors with anti-deaminases derived from viruses
Nature Communications (2022)
-
PROTAC targeted protein degraders: the past is prologue
Nature Reviews Drug Discovery (2022)
-
CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation
Nature Chemical Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.