Abstract
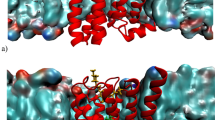
Despite recent advances in crystallography and the availability of G-protein-coupled receptor (GPCR) structures, little is known about the mechanism of their activation process, as only the β2 adrenergic receptor (β2AR) and rhodopsin have been crystallized in fully active conformations. Here we report the structure of an agonist-bound, active state of the human M2 muscarinic acetylcholine receptor stabilized by a G-protein mimetic camelid antibody fragment isolated by conformational selection using yeast surface display. In addition to the expected changes in the intracellular surface, the structure reveals larger conformational changes in the extracellular region and orthosteric binding site than observed in the active states of the β2AR and rhodopsin. We also report the structure of the M2 receptor simultaneously bound to the orthosteric agonist iperoxo and the positive allosteric modulator LY2119620. This structure reveals that LY2119620 recognizes a largely pre-formed binding site in the extracellular vestibule of the iperoxo-bound receptor, inducing a slight contraction of this outer binding pocket. These structures offer important insights into the activation mechanism and allosteric modulation of muscarinic receptors.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Accessions
Protein Data Bank
Data deposits
Coordinates and structure factors for the active M2 receptor in complex with Nb9-8 and iperoxo are deposited in the Protein Data Bank under accession code 4MQS, and the coordinates and structure factors of the same complex bound additionally to the allosteric modulator LY2119620 are deposited under accession code 4MQT.
References
Wess, J., Eglen, R. M. & Gautam, D. Muscarinic acetylcholine receptors: mutant mice provide new insights for drug development. Nature Rev. Drug Discov. 6, 721–733 (2007)
Peterson, G. L., Herron, G. S., Yamaki, M., Fullerton, D. S. & Schimerlik, M. I. Purification of the muscarinic acetylcholine receptor from porcine atria. Proc. Natl Acad. Sci. USA 81, 4993–4997 (1984)
Kubo, T. et al. Primary structure of porcine cardiac muscarinic acetylcholine receptor deduced from the cDNA sequence. FEBS Lett. 209, 367–372 (1986)
Mohr, K., Trankle, C. & Holzgrabe, U. Structure/activity relationships of M2 muscarinic allosteric modulators. Receptors Channels 9, 229–240 (2003)
Digby, G. J., Shirey, J. K. & Conn, P. J. Allosteric activators of muscarinic receptors as novel approaches for treatment of CNS disorders. Mol. Biosyst. 6, 1345–1354 (2010)
Keov, P., Sexton, P. M. & Christopoulos, A. Allosteric modulation of G protein-coupled receptors: a pharmacological perspective. Neuropharmacology 60, 24–35 (2011)
Haga, K. et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature 482, 547–551 (2012)
Kruse, A. C. et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature 482, 552–556 (2012)
Choe, H. W. et al. Crystal structure of metarhodopsin II. Nature 471, 651–655 (2011)
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011)
Rasmussen, S. G. et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 (2011)
Deupi, X. et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc. Natl Acad. Sci. USA 109, 119–124 (2012)
Scheerer, P. et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 (2008)
Nygaard, R. et al. The dynamic process of β2-adrenergic receptor activation. Cell 152, 532–542 (2013)
Kloeckner, J., Schmitz, J. & Holzgrabe, U. Convergent, short synthesis of the muscarinic superagonist iperoxo. Tetrahedr. Lett. 51, 3470–3472 (2010)
Hudgins, P. M. & Stubbins, J. F. A comparison of the action of acetylcholine and acetylcholine mustard (chloroethylmethylaminoethyl acetate) on muscarinic and nicotinic receptors. J. Pharmacol. Exp. Ther. 182, 303–311 (1972)
Spalding, T. A., Birdsall, N. J., Curtis, C. A. & Hulme, E. C. Acetylcholine mustard labels the binding site aspartate in muscarinic acetylcholine receptors. J. Biol. Chem. 269, 4092–4097 (1994)
Ring, A. M. et al. Adrenaline-activated structure of the β2-adrenoceptor stabilized by an engineered nanobody. Nature 502, 575–579 (2013)
Miao, Y., Nichols, S. E., Gasper, P. M., Metzger, V. T. & McCammon, J. A. Activation and dynamic network of the M2 muscarinic receptor. Proc. Natl Acad. Sci. USA 110, 10982–10987 (2013)
Ballesteros, J. A. et al. Activation of the β2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J. Biol. Chem. 276, 29171–29177 (2001)
Heitz, F. et al. Site-directed mutagenesis of the putative human muscarinic M2 receptor binding site. Eur. J. Pharmacol. 380, 183–195 (1999)
Wess, J., Maggio, R., Palmer, J. R. & Vogel, Z. Role of conserved threonine and tyrosine residues in acetylcholine binding and muscarinic receptor activation. A study with m3 muscarinic receptor point mutants. J. Biol. Chem. 267, 19313–19319 (1992)
Vogel, W. K., Sheehan, D. M. & Schimerlik, M. I. Site-directed mutagenesis on the m2 muscarinic acetylcholine receptor: the significance of Tyr 403 in the binding of agonists and functional coupling. Mol. Pharmacol. 52, 1087–1094 (1997)
Gregory, K. J., Hall, N. E., Tobin, A. B., Sexton, P. M. & Christopoulos, A. Identification of orthosteric and allosteric site mutations in M2 muscarinic acetylcholine receptors that contribute to ligand-selective signaling bias. J. Biol. Chem. 285, 7459–7474 (2010)
De Amici, M., Dallanoce, C., Holzgrabe, U., Trankle, C. & Mohr, K. Allosteric ligands for G protein-coupled receptors: a novel strategy with attractive therapeutic opportunities. Med. Res. Rev. 30, 463–549 (2010)
Gregory, K. J., Sexton, P. M. & Christopoulos, A. Allosteric modulation of muscarinic acetylcholine receptors. Curr. Neuropharmacol. 5, 157–167 (2007)
Bock, A. et al. The allosteric vestibule of a seven transmembrane helical receptor controls G-protein coupling. Nature Commun. 3, 1044 (2012)
May, L. T. et al. Structure-function studies of allosteric agonism at M2 muscarinic acetylcholine receptors. Mol. Pharmacol. 72, 463–476 (2007)
Valant, C., Felder, C. C., Sexton, P. M. & Christopoulos, A. Probe dependence in the allosteric modulation of a G protein-coupled receptor: implications for detection and validation of allosteric ligand effects. Mol. Pharmacol. 81, 41–52 (2012)
Prilla, S., Schrobang, J., Ellis, J., Holtje, H. D. & Mohr, K. Allosteric interactions with muscarinic acetylcholine receptors: complex role of the conserved tryptophan M2422Trp in a critical cluster of amino acids for baseline affinity, subtype selectivity, and cooperativity. Mol. Pharmacol. 70, 181–193 (2006)
Chee, M. J. et al. The third intracellular loop stabilizes the inactive state of the neuropeptide Y1 receptor. J. Biol. Chem. 283, 33337–33346 (2008)
Broach, J. R. & Thorner, J. High-throughput screening for drug discovery. Nature 384, 14–16 (1996)
Ehlert, F. J. Estimation of the affinities of allosteric ligands using radioligand binding and pharmacological null methods. Mol. Pharmacol. 33, 187–194 (1988)
Canals, M. et al. A Monod-Wyman-Changeux mechanism can explain G protein-coupled receptor (GPCR) allosteric modulation. J. Biol. Chem. 287, 650–659 (2012)
Leach, K., Sexton, P. M. & Christopoulos, A. Allosteric GPCR modulators: taking advantage of permissive receptor pharmacology. Trends Pharmacol. Sci. 28, 382–389 (2007)
Ichiyama, S. et al. The structure of the third intracellular loop of the muscarinic acetylcholine receptor M2 subtype. FEBS Lett. 580, 23–26 (2006)
Shapiro, R. A. & Nathanson, N. M. Deletion analysis of the mouse m1 muscarinic acetylcholine receptor: effects on phosphoinositide metabolism and down-regulation. Biochemistry 28, 8946–8950 (1989)
Conrath, K. E. et al. β-Lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob. Agents Chemother. 45, 2807–2812 (2001)
Whorton, M. R. et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl Acad. Sci. USA 104, 7682–7687 (2007)
Hu, J. et al. Structural basis of G protein-coupled receptor-G protein interactions. Nature Chem. Biol. 6, 541–548 (2010)
Liu, J., Conklin, B. R., Blin, N., Yun, J. & Wess, J. Identification of a receptor/G-protein contact site critical for signaling specificity and G-protein activation. Proc. Natl Acad. Sci. USA 92, 11642–11646 (1995)
Li, B. et al. Rapid identification of functionally critical amino acids in a G protein-coupled receptor. Nature Methods 4, 169–174 (2007)
McMillin, S. M., Heusel, M., Liu, T., Costanzi, S. & Wess, J. Structural basis of M3 muscarinic receptor dimer/oligomer formation. J. Biol. Chem. 286, 28584–28598 (2011)
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nature Protocols 4, 706–731 (2009)
Otwinowski, Z. & Minor, W. in Methods in Enzymology Vol. 276 (ed. Carter, C. W. ) 307–326 (Academic, 1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Acknowledgements
We acknowledge support from the National Science Foundation (graduate fellowship to A.C.K., and Award 1223785 to B.K.K.), the Stanford Medical Scientist Training Program (A.M. and A.M.R.), the American Heart Association (A.M.), the Ruth L. Kirschstein National Research Service Award (A.M.R.), National Institutes of Health grants NS02847123 and GM08311806 (B.K.K.), the Mathers Foundation (B.K.K., W.I.W. and K.C.G.), the Deutsche Forschungsgemeinschaft for the grant GM 13/10-1 (K.E., H.H., P.G.), the National Health and Medical Research Council (NHMRC) of Australia program grant 519461 (P.M.S. and A.C.), NHMRC Principal Research Fellowships (P.M.S. and A.C.), and the Howard Hughes Medical Institute (K.C.G.). This work was supported in part by the Intramural Research Program, NIDDK, NIH, US Department of Health and Human Services (J.H., K.H. and J.W.). We thank K. Leach for performing ERK assays, and B. Davie and P. Scammells for synthesis of iperoxo. We thank H. Xiao, C. H. Croy and D. A. Schober for functional characterization of LY2119620. We thank T. S. Kobilka for preparation of affinity chromatography reagents and F. S. Thian for help with cell culture.
Author information
Authors and Affiliations
Contributions
A.C.K. expressed and purified M2 receptor for yeast display and crystallographic experiments, performed crystallization, data collection, and structure refinement, and performed radioligand binding assays to validate nanobody activity. A.C.K., A.M.R. and A.M. designed experiments to identify nanobodies by yeast display. A.M.R. performed all yeast selections, and expressed and purified Nb9-8 and other nanobodies. J.H. and K.H. performed site-directed mutagenesis and characterization of resulting mutants. K.E. synthesized FAUC123. H.H. performed cell assays and radioligand binding to characterize FAUC123. C.V. performed pharmacological characterization of LY2119620. P.M.S. and A.C. supervised pharmacological characterization of LY2119620. C.C.F. designed key solubility, physical chemistry and ligand analysis to select LY2119620 as an appropriate co-crystallization candidate for the M2 receptor. P.G. supervised synthesis and characterization of FAUC123. E.P. and J.S. performed llama immunization, cDNA production, and performed selections by phage display. W.I.W. supervised structure refinement. K.C.G. supervised yeast selection experiments. J.W. supervised mutagenesis experiments and analysed results. B.K.K. provided overall project supervision, and with A.C.K., A.M.R. and A.M. wrote the manuscript with assistance from A.C. and J.W.
Corresponding author
Ethics declarations
Competing interests
A.C.K., A.M.R. and A.M. have applied for a patent on the yeast display and screening methods used to identify the conformationally selective nanobody used to obtain the crystal structure.
Extended data figures and tables
Extended Data Figure 1 Characterization of FAUC123.
a, Activation of M2 receptor by the prototypical muscarinic agonist carbachol, the high-affinity agonist iperoxo, and an irreversible iperoxo analogue (FAUC123) shows that iperoxo and FAUC123 are exceptionally potent full agonists at the M2 muscarinic receptor. Points indicate mean ± s.e.m. of three independent measurements, each performed in triplicate. b, Sf9 membranes expressing the human M2 receptor were incubated overnight at 4 °C with either no ligand, 100 μM iperoxo, or 100 μM FAUC123. Membranes were then washed three times in buffer without ligand, and incubated with a saturating concentration (20 nM) of [3H]-NMS. Incubation with iperoxo had no effect on radioligand binding, whereas FAUC123 blocked almost all [3H]-NMS binding sites. Bars indicate mean ± s.e.m. of three independent measurements. c, FAUC123 was tested for its ability to induce M2 receptor activation after covalent modification. Whereas iperoxo-induced inositol phosphate production was blocked by 1 μM atropine, FAUC123-induced activation was not susceptible to atropine blockade. Bars indicate mean ± s.e.m. of three independent measurements.
Extended Data Figure 2 Comparison to other active GPCR structures.
Structures of all activated GPCRs show similarities in conformational changes at the intracellular surface. In each case, the intracellular tip of transmembrane helix 6 (TM6) moves outward on activation, as seen in the view from the intracellular side (right panels). This creates a cavity to which a G protein can bind the receptor.
Extended Data Figure 3 Pharmacology.
a, Functional properties are shown for M2 receptors in which key residues were mutated. Agonist-induced increases in intracellular calcium levels were monitored via FLIPR using transfected COS-7 cells. Because some mutant receptors (N58A, D103E) were expressed at lower levels than the wild-type (WT) receptor, reference curves were obtained using cells transfected with either 3 μg DNA or 1 μg wild-type receptor DNA. The latter cells showed receptor expression levels comparable to those found with the N58A and D103E mutants (see Extended Data Table 2 for details). Data are given as means ± s.e.m. of three independent experiments, each carried out in triplicate. AU, arbitrary units. b, The interaction between LY2119620 and iperoxo was measured by radioligand binding and functional assays. LY2119620 enhances the affinity of iperoxo (top graph) and its signalling potency (bottom graphs), and is also able to activate M2 receptor signalling directly as measured by [35S]GTPγS and ERK1/2 phosphorylation. Experiments were carried out with CHO cells stably expressing the human M2 receptor, and points are shown as mean ± s.e.m. of three independent experiments, each carried out in duplicate.
Extended Data Figure 4 Binding-site diagram.
M2 receptor residues interacting with the orthosteric agonist iperoxo and the positive allosteric modulator LY2119620 are shown. Polar contacts are highlighted as red dotted lines, and hydrophobic contacts are in green solid lines.
Extended Data Figure 5 Electron density.
a, b, Fo − Fc omit maps are shown in grey, contoured at 2.5σ within a 2.5 Å radius of the indicated ligand. c–f, 2Fo − Fc maps are shown in blue, contoured at 1.5σ within a 2.0 Å radius of the indicated region.
Extended Data Figure 6 Comparison of M2 receptor structures with and without LY2119620 bound.
Comparison of the structure of active M2 receptor with and without the allosteric modulator LY2119620 reveals that there are few differences outside the extracellular vestibule. The overall structures are compared in a. Within the extracellular vestibule, there is a slight contraction in the presence of the modulator, and Trp 4227.35 undergoes a change of rotamer (panel b, red arrow). The orthosteric ligand-binding site, c, and intracellular surface, d, show few differences.
Supplementary information
Supplementary Information
This file contains supplementary text and data and additional references. (PDF 494 kb)
Rights and permissions
About this article
Cite this article
Kruse, A., Ring, A., Manglik, A. et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106 (2013). https://doi.org/10.1038/nature12735
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12735
This article is cited by
-
Design of supramolecular nanosheets for drug delivery applications
Polymer Journal (2023)
-
Activation and signaling mechanism revealed by GPR119-Gs complex structures
Nature Communications (2022)
-
Reconstitution of microtubule into GTP-responsive nanocapsules
Nature Communications (2022)
-
Pushing the frontiers: tools for monitoring neurotransmitters and neuromodulators
Nature Reviews Neuroscience (2022)
-
Selective targeting of ligand-dependent and -independent signaling by GPCR conformation-specific anti-US28 intrabodies
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.