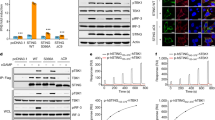
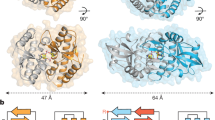
Abstract
Cytosolic DNA arising from intracellular bacterial or viral infections is a powerful pathogen-associated molecular pattern (PAMP) that leads to innate immune host defence by the production of type I interferon and inflammatory cytokines. Recognition of cytosolic DNA by the recently discovered cyclic-GMP-AMP (cGAMP) synthase (cGAS) induces the production of cGAMP to activate the stimulator of interferon genes (STING). Here we report the crystal structure of cGAS alone and in complex with DNA, ATP and GTP along with functional studies. Our results explain the broad DNA sensing specificity of cGAS, show how cGAS catalyses dinucleotide formation and indicate activation by a DNA-induced structural switch. cGAS possesses a remarkable structural similarity to the antiviral cytosolic double-stranded RNA sensor 2′-5′oligoadenylate synthase (OAS1), but contains a unique zinc thumb that recognizes B-form double-stranded DNA. Our results mechanistically unify dsRNA and dsDNA innate immune sensing by OAS1 and cGAS nucleotidyl transferases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Rathinam, V. A. K. & Fitzgerald, K. A. Cytosolic surveillance and antiviral immunity. Curr. Opin. Virol. 1, 455–462 (2011)
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010)
Keating, S. E., Baran, M. & Bowie, A. G. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 32, 574–581 (2011)
Krug, A. Nucleic acid recognition receptors in autoimmunity. Handb. Exp. Pharmacol. 183, 129–151 (2008)
Takaoka, A. et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature 448, 501–505 (2007)
Bürckstümmer, T. et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nature Immunol. 10, 266–272 (2009)
Fernandes-Alnemri, T., Yu, J. W., Datta, P., Wu, J. & Alnemri, E. S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 (2009)
Hornung, V. et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 (2009)
Ablasser, A. et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nature Immunol. 10, 1065–1072 (2009)
Chiu, Y. H., Macmillan, J. B. & Chen, Z. J. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138, 576–591 (2009)
Yang, P. et al. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a β-catenin-dependent pathway. Nature Immunol. 11, 487–494 (2010)
Kim, T. et al. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc. Natl Acad. Sci. USA 107, 15181–15186 (2010)
Zhang, Z. et al. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature Immunol. 12, 959–965 (2011)
Unterholzner, L. et al. IFI16 is an innate immune sensor for intracellular DNA. Nature Immunol. 11, 997–1004 (2010)
Rathinam, V. A. & Fitzgerald, K. A. Innate immune sensing of DNA viruses. Virology 411, 153–162 (2011)
Ishikawa, H., Ma, Z. & Barber, G. N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792 (2009)
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008)
Wu, J. et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013)
Abe, T. et al. STING recognition of cytoplasmic DNA instigates cellular defense. Mol. Cell 50, 5–15 (2013)
Burdette, D. L. et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011)
McWhirter, S. M. et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 206, 1899–1911 (2009)
Woodward, J. J., Iavarone, A. T. & Portnoy, D. A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705 (2010)
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013)
Kuchta, K., Knizewski, L., Wyrwicz, L. S., Rychlewski, L. & Ginalski, K. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res. 37, 7701–7714 (2009)
Cui, S. et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29, 169–179 (2008)
Witte, G., Hartung, S., Buttner, K. & Hopfner, K. P. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell 30, 167–178 (2008)
Stagno, J., Aphasizheva, I., Rosengarth, A., Luecke, H. & Aphasizhev, R. UTP-bound and apo structures of a minimal RNA uridylyltransferase. J. Mol. Biol. 366, 882–899 (2007)
Xiong, Y. & Steitz, T. A. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature 430, 640–645 (2004)
Ablasser, A. et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING Nature http://dx.doi.org/10.1038/nature12306 (30 May 2013)
Hartmann, R., Justesen, J., Sarkar, S. N., Sen, G. C. & Yee, V. C. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol. Cell 12, 1173–1185 (2003)
Donovan, J., Dufner, M. & Korennykh, A. Structural basis for cytosolic double-stranded RNA surveillance by human oligoadenylate synthetase 1. Proc. Natl Acad. Sci. USA 110, 1652–1657 (2013)
Gao, P. et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107 (2013)
Cavlar, T., Deimling, T., Ablasser, A., Hopfner, K. P. & Hornung, V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 32, 1440–1450 (2013)
Kabsch, W. XDS. Acta Crystallogr. 66, 125–132 (2010)
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007)
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. 62, 1002–1011 (2006)
Morris, R. J., Perrakis, A. & Lamzin, V. S. ARP/wARP’s model-building algorithms. I. The main chain. Acta Crystallogr. 58, 968–975 (2002)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 66, 213–221 (2010)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. 60, 2210–2221 (2004)
Rao, F. et al. Enzymatic synthesis of c-di-GMP using a thermophilic diguanylate cyclase. Anal. Biochem. 389, 138–142 (2009)
Acknowledgements
We thank A. Butryn for comments on the manuscript. We thank the Max-Planck-Crystallization facility for initial crystal screening and the Swiss Light Source, European Synchrotron Radiation Facility and the German electron synchrotron Petra III for beam time and on-site assistance. This work was funded by the National Institutes of Health (U19AI083025), the European Research Council Advanced Grant 322869, and the Center for Integrated Protein Science Munich (CIPSM) to K.-P.H., by DFG grant 3717/2-1 to G.W., by GRK1721 to K.-P.H. and G.W., by DFG grant SFB670 and ERC grant 243046 to V.H.; C.C.O.M. is supported by GRK1721.
Author information
Authors and Affiliations
Contributions
F.C. crystallized and determined the structure of cGAS, performed biochemical assays, interpreted data and wrote the manuscript. T.D. crystallized and refined the DNA complex. C.C.O.M., A.A., T.D. and M.M. performed biochemical assays. G.W. performed biochemical assays, interpreted data and helped with structure determination. V.H. supervised the cell-based experiments and interpreted data. K.-P.H designed the research, helped with structure determination, interpreted data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1–6 and Supplementary Table 1. The figures show the activity both ‘in vitro’ and ‘in cells’, sequence conservation and secondary structure and the table summarizes the crystallographic data collection and model refinement statistics. (PDF 4253 kb)
Rights and permissions
About this article
Cite this article
Civril, F., Deimling, T., de Oliveira Mann, C. et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337 (2013). https://doi.org/10.1038/nature12305
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12305
This article is cited by
-
Cytosolic DNA sensors in neurodegenerative diseases: from physiological defenders to pathological culprits
EMBO Molecular Medicine (2024)
-
Second messenger 2'3'-cyclic GMP-AMP (2'3'-cGAMP): the cell autonomous and non-autonomous roles in cancer progression
Acta Pharmacologica Sinica (2024)
-
MRE11 liberates cGAS from nucleosome sequestration during tumorigenesis
Nature (2024)
-
Biological parameters for quality evaluation of allografts from the Brazilian National Institute of Traumatology and Orthopedics tissue bank
Cell and Tissue Banking (2024)
-
Candida albicans extracellular vesicles trigger type I IFN signalling via cGAS and STING
Nature Microbiology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.